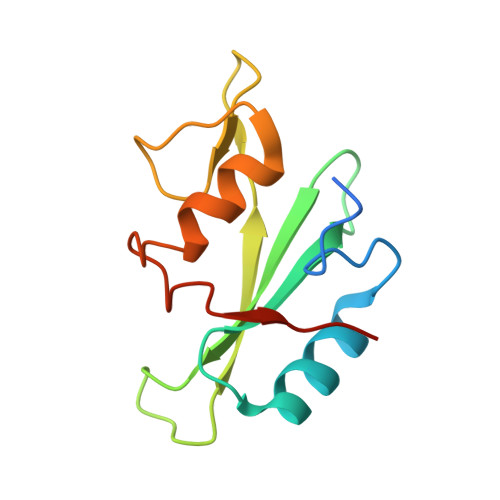
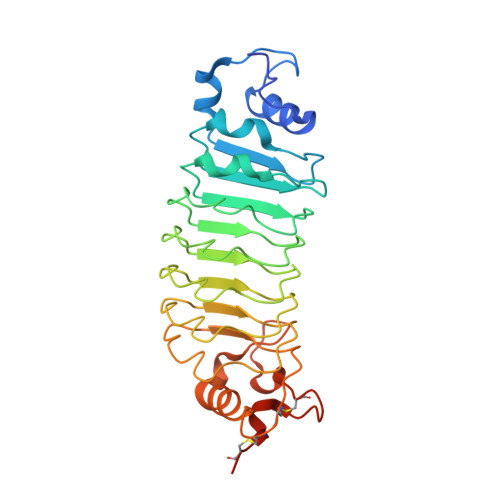
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms.
Duarte, D.P., Lamontanara, A.J., La Sala, G., Jeong, S., Sohn, Y.K., Panjkovich, A., Georgeon, S., Kukenshoner, T., Marcaida, M.J., Pojer, F., De Vivo, M., Svergun, D., Kim, H.S., Dal Peraro, M., Hantschel, O.(2020) Nat Commun 11: 2319-2319
- PubMed: 32385234
- DOI: https://doi.org/10.1038/s41467-020-16128-5
- Primary Citation of Related Structures:
6HTF - PubMed Abstract:
Bruton's tyrosine kinase (Btk) is critical for B-cell maturation and activation. Btk loss-of-function mutations cause human X-linked agammaglobulinemia (XLA). In contrast, Btk signaling sustains growth of several B-cell neoplasms which may be treated with tyrosine kinase inhibitors (TKIs). Here, we uncovered the structural mechanism by which certain XLA mutations in the SH2 domain strongly perturb Btk activation. Using a combination of molecular dynamics (MD) simulations and small-angle X-ray scattering (SAXS), we discovered an allosteric interface between the SH2 and kinase domain required for Btk activation and to which multiple XLA mutations map. As allosteric interactions provide unique targeting opportunities, we developed an engineered repebody protein binding to the SH2 domain and able to disrupt the SH2-kinase interaction. The repebody prevents activation of wild-type and TKI-resistant Btk, inhibiting Btk-dependent signaling and proliferation of malignant B-cells. Therefore, the SH2-kinase interface is critical for Btk activation and a targetable site for allosteric inhibition.
- Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, École polytechnique fédérale de Lausanne (EPFL, 1015, Lausanne, Switzerland.
Organizational Affiliation: