RXRa structure complexed with CU-6PMN and SRC1 peptide.
Kawasaki, M., Nakano, S., Motoyama, T., Yamada, S., Watanabe, M., Takamura, Y., Fujihara, M., Tokiwa, H., Kakuta, H., Ito, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Retinoic acid receptor RXR-alpha | 243 | Homo sapiens | Mutation(s): 0 Gene Names: RXRA, NR2B1 | 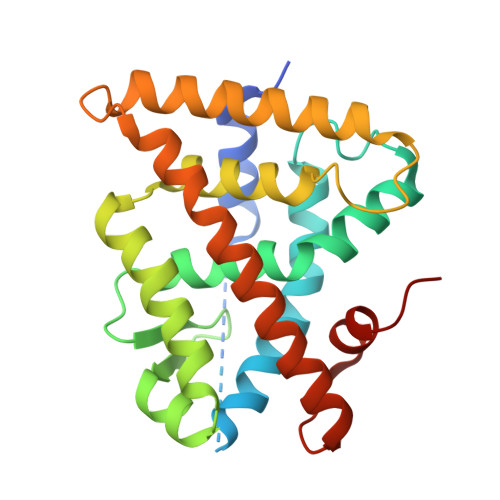 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P19793 (Homo sapiens) Explore P19793 Go to UniProtKB: P19793 | |||||
PHAROS: P19793 GTEx: ENSG00000186350 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19793 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN | 12 | Homo sapiens | Mutation(s): 0 EC: 2.3.1.48 | 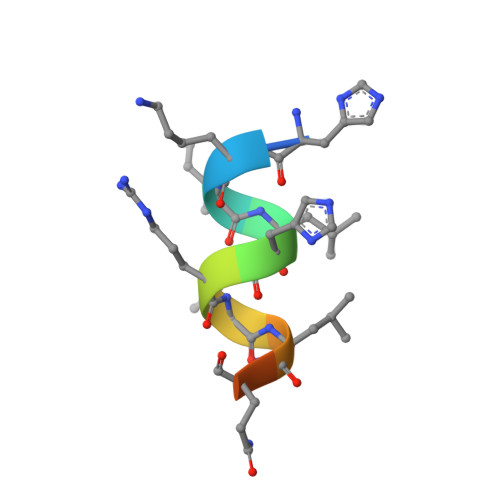 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15788 (Homo sapiens) Explore Q15788 Go to UniProtKB: Q15788 | |||||
PHAROS: Q15788 GTEx: ENSG00000084676 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15788 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| WY5 (Subject of Investigation/LOI) Query on WY5 | E [auth A], F [auth B] | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid C25 H26 O5 UYPOSIGCFZMVDD-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 64.39 | α = 90 |
| b = 64.39 | β = 90 |
| c = 111.168 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science | Japan | 16K18688 |
| Japan Society for the Promotion of Science | Japan | 18K14391 |
| Japan Society for the Promotion of Science | Japan | 12J06716 |