LSD1/Co-Rest structure with an inhibitor
Wang, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lysine-specific histone demethylase 1A | 662 | Homo sapiens | Mutation(s): 0 Gene Names: KDM1A, AOF2, KDM1, KIAA0601, LSD1 EC: 1 (PDB Primary Data), 1.14.11 (UniProt), 1.14.99.66 (UniProt), 1.14.11.65 (UniProt) | 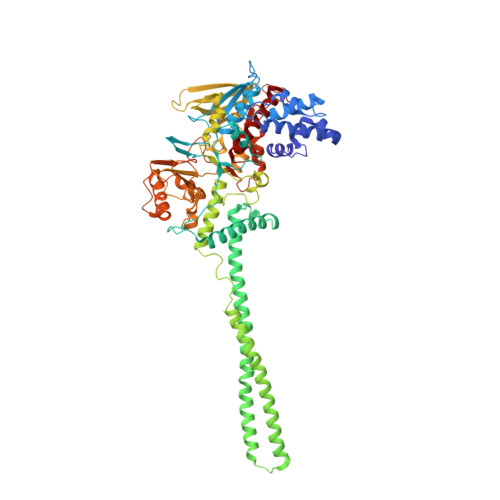 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O60341 (Homo sapiens) Explore O60341 Go to UniProtKB: O60341 | |||||
PHAROS: O60341 GTEx: ENSG00000004487 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60341 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REST corepressor 1 | 140 | Homo sapiens | Mutation(s): 0 Gene Names: RCOR1, KIAA0071, RCOR | 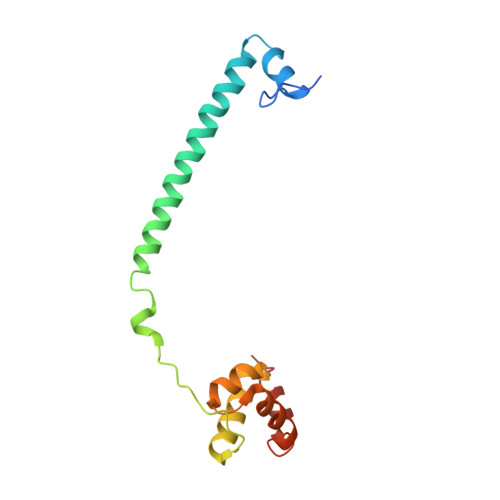 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UKL0 (Homo sapiens) Explore Q9UKL0 Go to UniProtKB: Q9UKL0 | |||||
PHAROS: Q9UKL0 GTEx: ENSG00000089902 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UKL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| D3U Query on D3U | N [auth A] | 2-PCPA derivative C36 H41 N9 O15 P2 LTYXJNFFNLDHFB-VXYXFEBYSA-N |  | ||
| CW0 (Subject of Investigation/LOI) Query on CW0 | C [auth A] | piperidin-4-ylmethyl 4-fluoranyl-4-[[[(1~{R},2~{S})-2-phenylcyclopropyl]amino]methyl]piperidine-1-carboxylate C22 H32 F N3 O2 CMYNDDNZDLGSJZ-VQTJNVASSA-N |  | ||
| DTT Query on DTT | D [auth A] | 2,3-DIHYDROXY-1,4-DITHIOBUTANE C4 H10 O2 S2 VHJLVAABSRFDPM-IMJSIDKUSA-N |  | ||
| PEG Query on PEG | AA [auth A], BA [auth A], CA [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| MLA Query on MLA | G [auth A], H [auth A], I [auth A] | MALONIC ACID C3 H4 O4 OFOBLEOULBTSOW-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | J [auth A], K [auth A], L [auth A], M [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | O [auth A] P [auth A] Q [auth A] R [auth A] S [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | E [auth A], F [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CL Query on CL | DA [auth A], EA [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 121.51 | α = 90 |
| b = 179.44 | β = 90 |
| c = 234.41 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data scaling |
| PHASER | phasing |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 81620108027 |