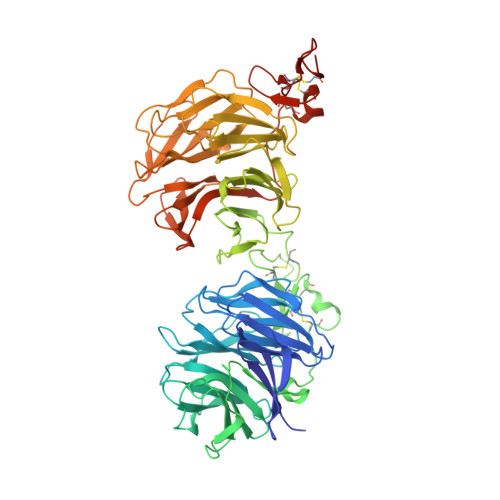
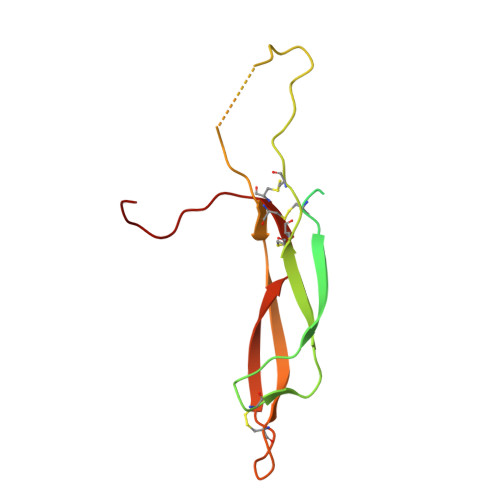
Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains.
Kim, J., Han, W., Park, T., Kim, E.J., Bang, I., Lee, H.S., Jeong, Y., Roh, K., Kim, J., Kim, J.S., Kang, C., Seok, C., Han, J.-K., Choi, H.-J.(2020) Nat Commun 11: 5357-5357
- PubMed: 33097721
- DOI: https://doi.org/10.1038/s41467-020-19155-4
- Primary Citation of Related Structures:
6L6R - PubMed Abstract:
Low-density lipoprotein receptor-related protein 6 (LRP6) is a coreceptor of the β-catenin-dependent Wnt signaling pathway. The LRP6 ectodomain binds Wnt proteins, as well as Wnt inhibitors such as sclerostin (SOST), which negatively regulates Wnt signaling in osteocytes. Although LRP6 ectodomain 1 (E1) is known to interact with SOST, several unresolved questions remain, such as the reason why SOST binds to LRP6 E1E2 with higher affinity than to the E1 domain alone. Here, we present the crystal structure of the LRP6 E1E2-SOST complex with two interaction sites in tandem. The unexpected additional binding site was identified between the C-terminus of SOST and the LRP6 E2 domain. This interaction was confirmed by in vitro binding and cell-based signaling assays. Its functional significance was further demonstrated in vivo using Xenopus laevis embryos. Our results provide insights into the inhibitory mechanism of SOST on Wnt signaling.
- Department of Biological Sciences, Seoul National University, Seoul, 08826, Republic of Korea.
Organizational Affiliation: