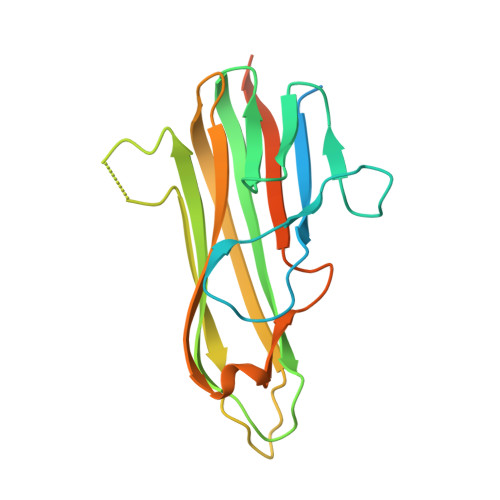
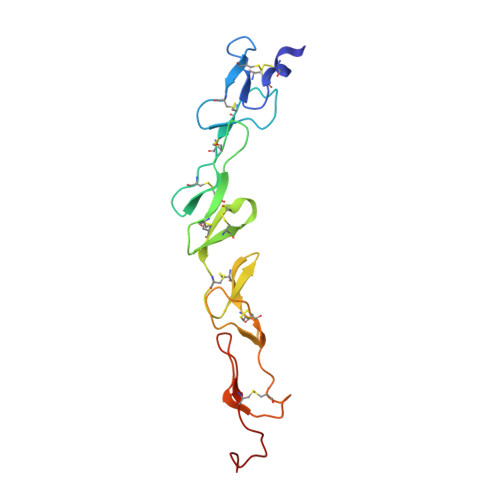
Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Kimberlin, C.R., Chin, S.M., Roe-Zurz, Z., Zhang, P., Xu, A., Liao-Chan, S., Debasish, S., Nager, A.R., Schirle Oakdale, N., Brown, C., Wang, F., Yang, Y., Lindquist, K., Yeung, Y.A., Salek-Ardakani, S., Chaparro-Riggers, J.(2018) Nat Commun 9: 4679-4691