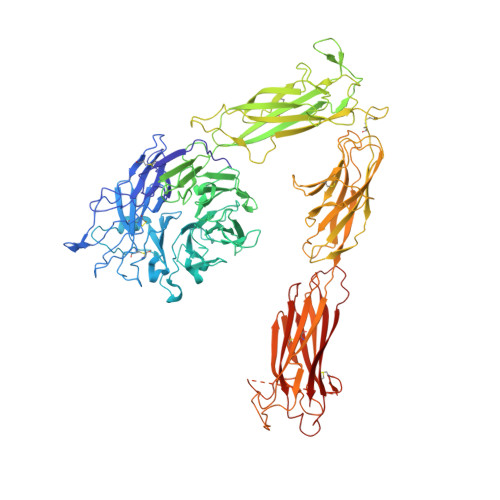
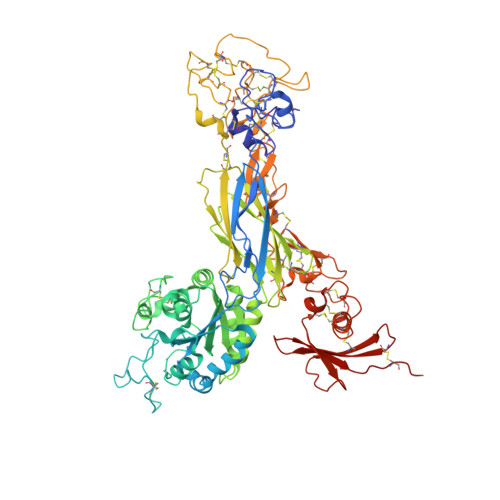
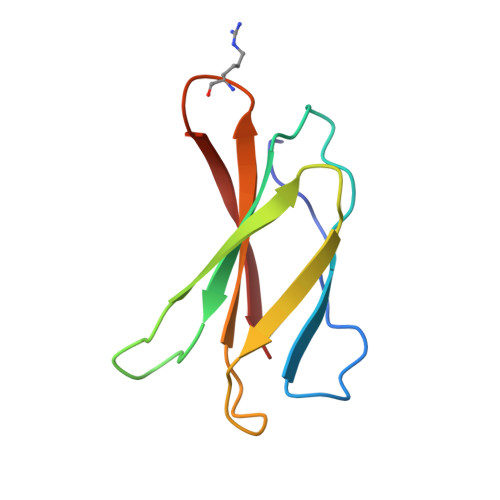
Structure-guided design of pure orthosteric inhibitors of alpha IIb beta 3 that prevent thrombosis but preserve hemostasis.
Adair, B.D., Alonso, J.L., van Agthoven, J., Hayes, V., Ahn, H.S., Yu, I.S., Lin, S.W., Xiong, J.P., Poncz, M., Arnaout, M.A.(2020) Nat Commun 11: 398-398
- PubMed: 31964886
- DOI: https://doi.org/10.1038/s41467-019-13928-2
- Primary Citation of Related Structures:
6NAJ - PubMed Abstract:
A prevailing dogma is that inhibition of vascular thrombosis by antagonizing platelet integrin αIIbβ3 cannot be achieved without compromising hemostasis, thus causing serious bleeding and increased morbidity and mortality. It is speculated that these adverse outcomes result from drug-induced activating conformational changes in αIIbβ3 but direct proof is lacking. Here, we report the structure-guided design of peptide Hr10 and a modified form of the partial agonist drug tirofiban that act as "pure" antagonists of αIIbβ3, i.e., they no longer induce the conformational changes in αIIbβ3. Both agents inhibit human platelet aggregation but preserve clot retraction. Hr10 and modified tirofiban are as effective as partial agonist drugs in inhibiting vascular thrombosis in humanized mice, but neither causes serious bleeding, establishing a causal link between partial agonism and impaired hemostasis. Pure orthosteric inhibitors of αIIbβ3 may thus provide safer alternatives for human therapy, and valuable tools to probe structure-activity relationships in integrins.
- Leukocyte Biology & Inflammation Program, and Structural Biology Program, Massachusetts General Hospital, Boston, MA, 02114, USA.
Organizational Affiliation: