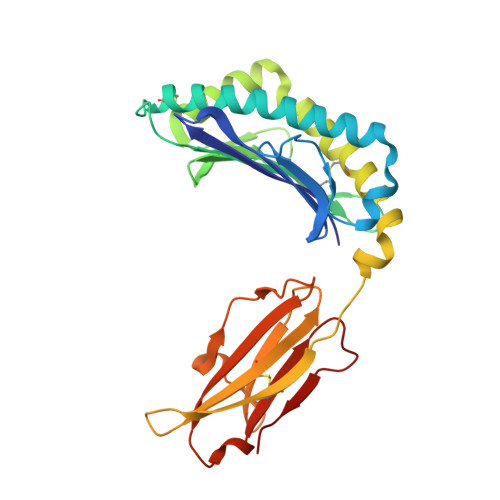
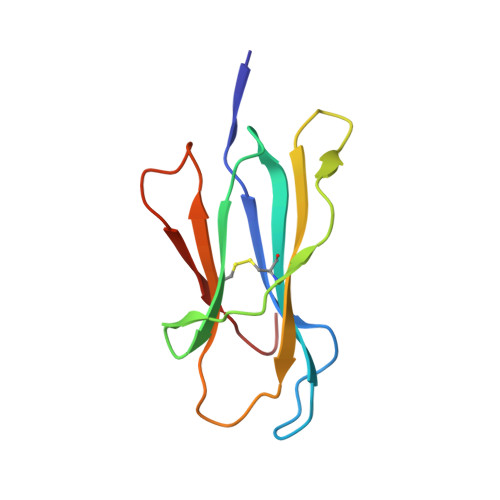
Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
McShan, A.C., Devlin, C.A., Overall, S.A., Park, J., Toor, J.S., Moschidi, D., Flores-Solis, D., Choi, H., Tripathi, S., Procko, E., Sgourakis, N.G.(2019) Proc Natl Acad Sci U S A 116: 25602-25613
- PubMed: 31796585
- DOI: https://doi.org/10.1073/pnas.1915562116
- Primary Citation of Related Structures:
6NPR - PubMed Abstract:
The interplay between a highly polymorphic set of MHC-I alleles and molecular chaperones shapes the repertoire of peptide antigens displayed on the cell surface for T cell surveillance. Here, we demonstrate that the molecular chaperone TAP-binding protein related (TAPBPR) associates with a broad range of partially folded MHC-I species inside the cell. Bimolecular fluorescence complementation and deep mutational scanning reveal that TAPBPR recognition is polarized toward the α 2 domain of the peptide-binding groove, and depends on the formation of a conserved MHC-I disulfide epitope in the α 2 domain. Conversely, thermodynamic measurements of TAPBPR binding for a representative set of properly conformed, peptide-loaded molecules suggest a narrower MHC-I specificity range. Using solution NMR, we find that the extent of dynamics at "hotspot" surfaces confers TAPBPR recognition of a sparsely populated MHC-I state attained through a global conformational change. Consistently, restriction of MHC-I groove plasticity through the introduction of a disulfide bond between the α 1 /α 2 helices abrogates TAPBPR binding, both in solution and on a cellular membrane, while intracellular binding is tolerant of many destabilizing MHC-I substitutions. Our data support parallel TAPBPR functions of 1) chaperoning unstable MHC-I molecules with broad allele-specificity at early stages of their folding process, and 2) editing the peptide cargo of properly conformed MHC-I molecules en route to the surface, which demonstrates a narrower specificity. Our results suggest that TAPBPR exploits localized structural adaptations, both near and distant to the peptide-binding groove, to selectively recognize discrete conformational states sampled by MHC-I alleles, toward editing the repertoire of displayed antigens.
- Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064.
Organizational Affiliation: