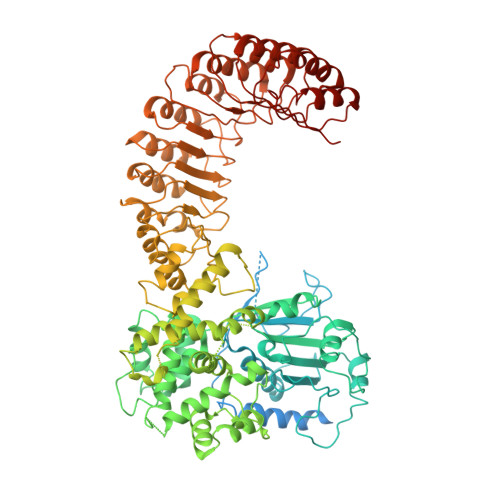
Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome.
Sharif, H., Wang, L., Wang, W.L., Magupalli, V.G., Andreeva, L., Qiao, Q., Hauenstein, A.V., Wu, Z., Nunez, G., Mao, Y., Wu, H.(2019) Nature 570: 338-343
- PubMed: 31189953
- DOI: https://doi.org/10.1038/s41586-019-1295-z
- Primary Citation of Related Structures:
6NPY - PubMed Abstract:
The NLRP3 inflammasome can be activated by stimuli that include nigericin, uric acid crystals, amyloid-β fibrils and extracellular ATP. The mitotic kinase NEK7 licenses the assembly and activation of the NLRP3 inflammasome in interphase. Here we report a cryo-electron microscopy structure of inactive human NLRP3 in complex with NEK7, at a resolution of 3.8 Å. The earring-shaped NLRP3 consists of curved leucine-rich-repeat and globular NACHT domains, and the C-terminal lobe of NEK7 nestles against both NLRP3 domains. Structural recognition between NLRP3 and NEK7 is confirmed by mutagenesis both in vitro and in cells. Modelling of an active NLRP3-NEK7 conformation based on the NLRC4 inflammasome predicts an additional contact between an NLRP3-bound NEK7 and a neighbouring NLRP3. Mutations to this interface abolish the ability of NEK7 or NLRP3 to rescue NLRP3 activation in NEK7-knockout or NLRP3-knockout cells. These data suggest that NEK7 bridges adjacent NLRP3 subunits with bipartite interactions to mediate the activation of the NLRP3 inflammasome.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: