A constricted opening in Kir channels does not impede potassium conduction.
Black, K.A., He, S., Jin, R., Miller, D.M., Bolla, J.R., Clarke, O.B., Johnson, P., Windley, M., Burns, C.J., Hill, A.P., Laver, D., Robinson, C.V., Smith, B.J., Gulbis, J.M.(2020) Nat Commun 11: 3024-3024
- PubMed: 32541684
- DOI: https://doi.org/10.1038/s41467-020-16842-0
- Primary Citation of Related Structures:
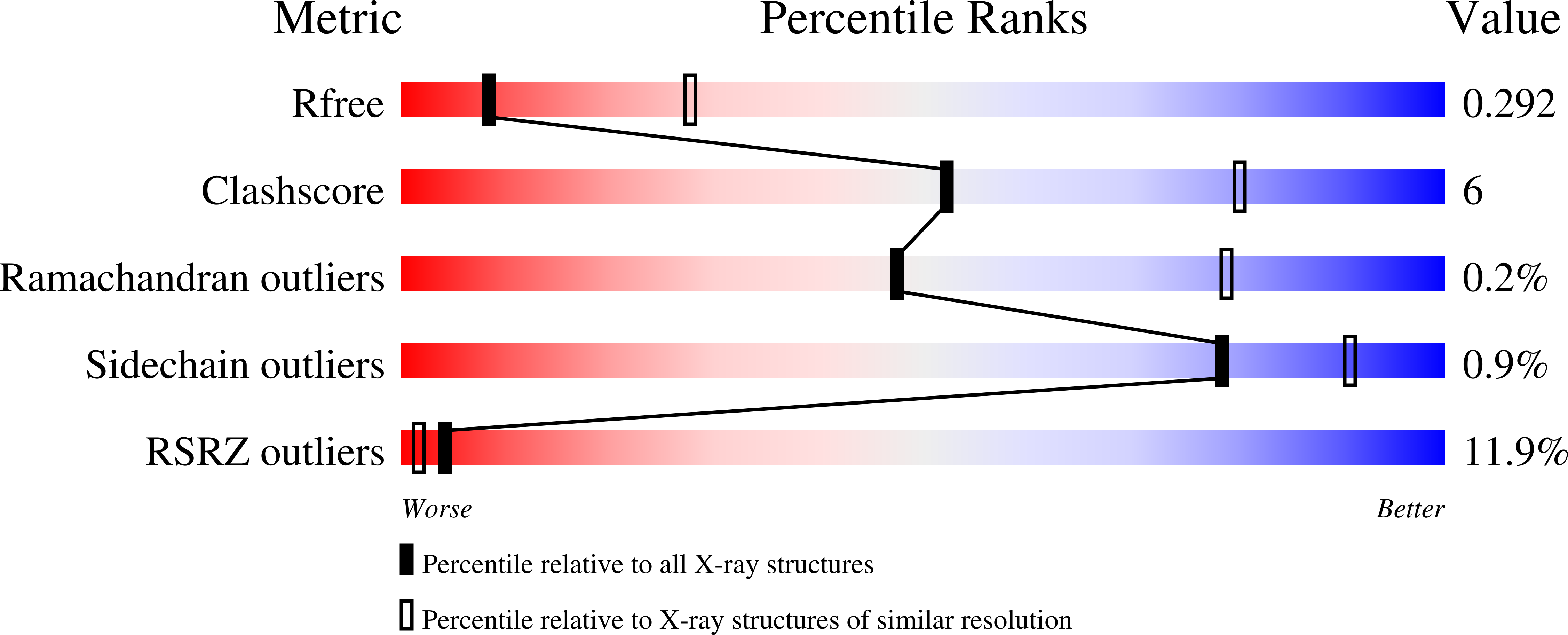
6O9T, 6O9U, 6O9V - PubMed Abstract:
The canonical mechanistic model explaining potassium channel gating is of a conformational change that alternately dilates and constricts a collar-like intracellular entrance to the pore. It is based on the premise that K + ions maintain a complete hydration shell while passing between the transmembrane cavity and cytosol, which must be accommodated. To put the canonical model to the test, we locked the conformation of a Kir K + channel to prevent widening of the narrow collar. Unexpectedly, conduction was unimpaired in the locked channels. In parallel, we employed all-atom molecular dynamics to simulate K + ions moving along the conduction pathway between the lower cavity and cytosol. During simulations, the constriction did not significantly widen. Instead, transient loss of some water molecules facilitated K + permeation through the collar. The low free energy barrier to partial dehydration in the absence of conformational change indicates Kir channels are not gated by the canonical mechanism.
Organizational Affiliation:
Structural Biology Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, 3052, Australia.