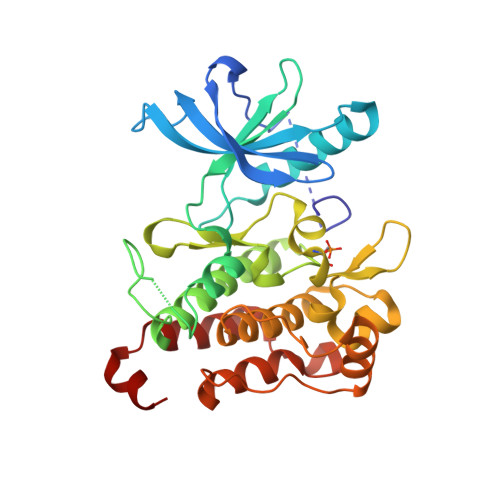
In Pursuit of an Allosteric Human Tropomyosin Kinase A (hTrkA) Inhibitor for Chronic Pain
Subramanian, G., Duclos, B., Johnson, P.D., Williams, T., Ross, J.T., Bowen, S.J., Zhu, Y., White, J.A., Hedke, C., Huczek, D., Collard, W., Javens, C., Vairagoundar, R., Respondek, T., Zachary, T., Maddux, T., Cox, M.R., Kamerling, S., Gonzales, A.J.(2021) ACS Med Chem Lett