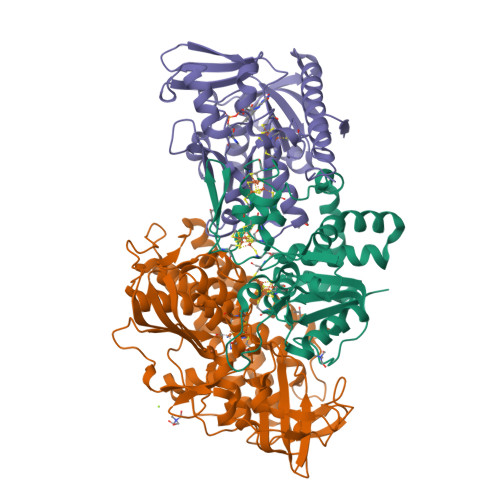
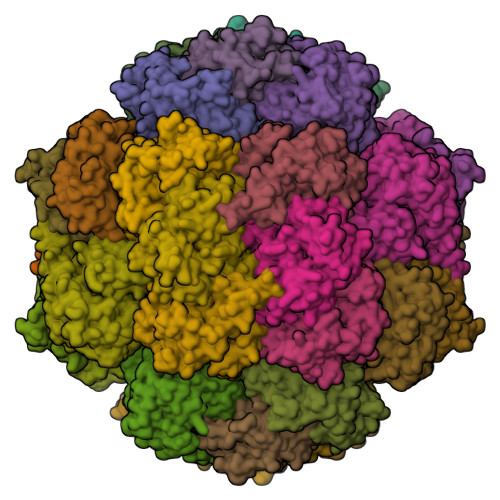
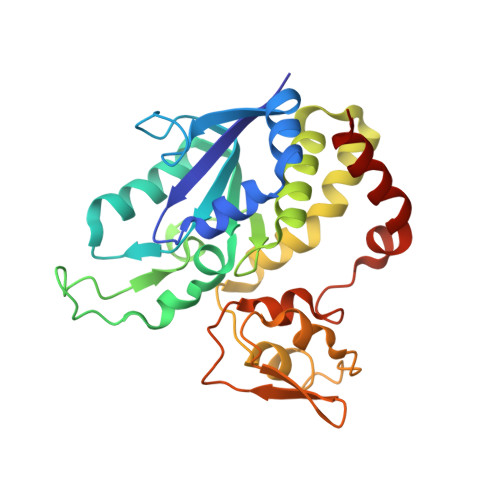
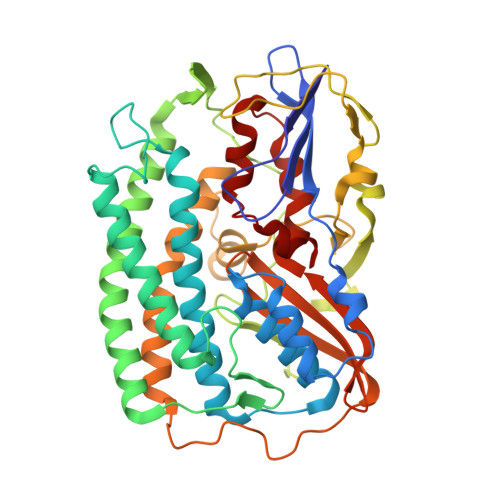
X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Ilina, Y., Lorent, C., Katz, S., Jeoung, J.H., Shima, S., Horch, M., Zebger, I., Dobbek, H.(2019) Angew Chem Int Ed Engl 58: 18710-18714
- PubMed: 31591784
- DOI: https://doi.org/10.1002/anie.201908258
- Primary Citation of Related Structures:
6QGR, 6QGT, 6QII - PubMed Abstract:
[NiFe] hydrogenases are complex model enzymes for the reversible cleavage of dihydrogen (H 2 ). However, structural determinants of efficient H 2 binding to their [NiFe] active site are not properly understood. Here, we present crystallographic and vibrational-spectroscopic insights into the unexplored structure of the H 2 -binding [NiFe] intermediate. Using an F 420 -reducing [NiFe]-hydrogenase from Methanosarcina barkeri as a model enzyme, we show that the protein backbone provides a strained chelating scaffold that tunes the [NiFe] active site for efficient H 2 binding and conversion. The protein matrix also directs H 2 diffusion to the [NiFe] site via two gas channels and allows the distribution of electrons between functional protomers through a subunit-bridging FeS cluster. Our findings emphasize the relevance of an atypical Ni coordination, thereby providing a blueprint for the design of bio-inspired H 2 -conversion catalysts.
Organizational Affiliation:
Institut für Biologie, Strukturbiologie/Biochemie, Humboldt-Universität zu Berlin, Philippstraße 13, 10115, Berlin, Germany.