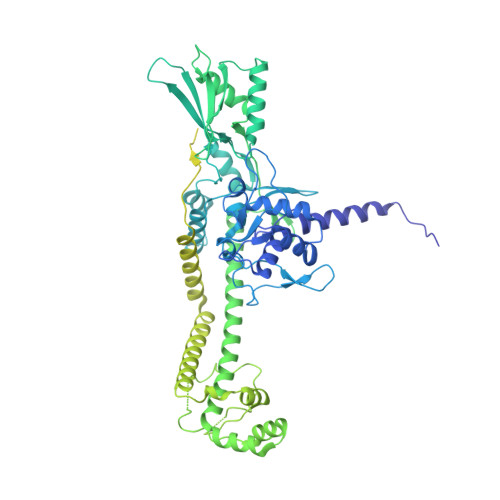
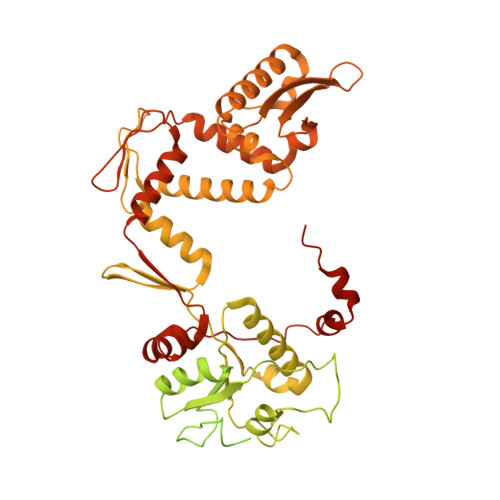
Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Vanden Broeck, A., Lotz, C., Ortiz, J., Lamour, V.(2019) Nat Commun 10: 4935-4935
- PubMed: 31666516
- DOI: https://doi.org/10.1038/s41467-019-12914-y
- Primary Citation of Related Structures:
6RKS, 6RKU, 6RKV, 6RKW - PubMed Abstract:
DNA gyrase is an essential enzyme involved in the homeostatic control of DNA supercoiling and the target of successful antibacterial compounds. Despite extensive studies, a detailed architecture of the full-length DNA gyrase from the model organism E. coli is still missing. Herein, we report the complete structure of the E. coli DNA gyrase nucleoprotein complex trapped by the antibiotic gepotidacin, using phase-plate single-particle cryo-electron microscopy. Our data unveil the structural and spatial organization of the functional domains, their connections and the position of the conserved GyrA-box motif. The deconvolution of two states of the DNA-binding/cleavage domain provides a better understanding of the allosteric movements of the enzyme complex. The local atomic resolution in the DNA-bound area reaching up to 3.0 Å enables the identification of the antibiotic density. Altogether, this study paves the way for the cryo-EM determination of gyrase complexes with antibiotics and opens perspectives for targeting conformational intermediates.
- Department of Integrated Structural Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), 1 Rue Laurent Fries, 67404, Illkirch Cedex, France.
Organizational Affiliation: