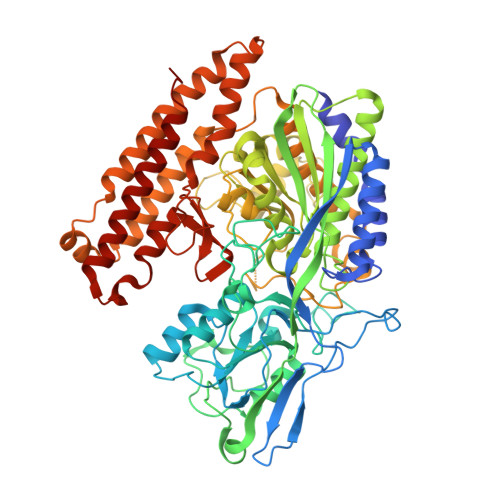
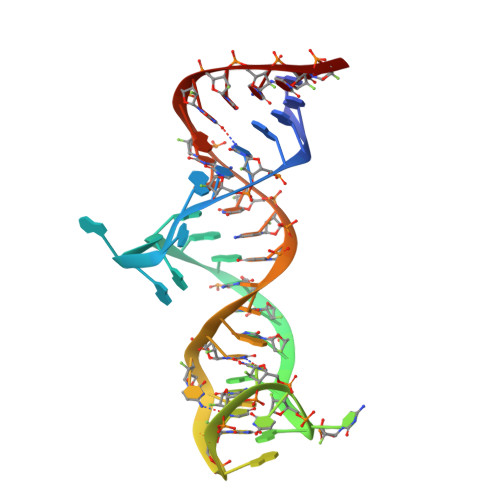
Structural basis of prostate-specific membrane antigen recognition by the A9g RNA aptamer.
Ptacek, J., Zhang, D., Qiu, L., Kruspe, S., Motlova, L., Kolenko, P., Novakova, Z., Shubham, S., Havlinova, B., Baranova, P., Chen, S.J., Zou, X., Giangrande, P., Barinka, C.(2020) Nucleic Acids Res 48: 11130-11145
- PubMed: 32525981
- DOI: https://doi.org/10.1093/nar/gkaa494
- Primary Citation of Related Structures:
6RTI - PubMed Abstract:
Prostate-specific membrane antigen (PSMA) is a well-characterized tumor marker associated with prostate cancer and neovasculature of most solid tumors. PSMA-specific ligands are thus being developed to deliver imaging or therapeutic agents to cancer cells. Here, we report on a crystal structure of human PSMA in complex with A9g, a 43-bp PSMA-specific RNA aptamer, that was determined to the 2.2 Å resolution limit. The analysis of the PSMA/aptamer interface allows for identification of key interactions critical for nanomolar binding affinity and high selectivity of A9g for human PSMA. Combined with in silico modeling, site-directed mutagenesis, inhibition experiments and cell-based assays, the structure also provides an insight into structural changes of the aptamer and PSMA upon complex formation, mechanistic explanation for inhibition of the PSMA enzymatic activity by A9g as well as its ligand-selective competition with small molecules targeting the internal pocket of the enzyme. Additionally, comparison with published protein-RNA aptamer structures pointed toward more general features governing protein-aptamer interactions. Finally, our findings can be exploited for the structure-assisted design of future A9g-based derivatives with improved binding and stability characteristics.
- Laboratory of Structural Biology, Institute of Biotechnology of the Czech Academy of Sciences, BIOCEV, Prumyslova 595, Vestec 25250, Czech Republic.
Organizational Affiliation: