Towards the mechanistic understanding of enzymatic CO2 reduction
Oliveira, A.R., Mota, C., Mourato, C., Domingos, R.M., Santos, M.F.A., Gesto, D., Guigliarelli, B., Santos-Silva, T., Romao, M.J., Pereira, I.C.(2020) ACS Catal
Experimental Data Snapshot
Starting Model: experimental
View more details
(2020) ACS Catal
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Formate dehydrogenase, alpha subunit, selenocysteine-containing,Formate dehydrogenase, alpha subunit, selenocysteine-containing,W-formate dehydrogenase - alpha subunit | 1,009 | Nitratidesulfovibrio vulgaris str. Hildenborough | Mutation(s): 0 EC: 1.2.1.2 | 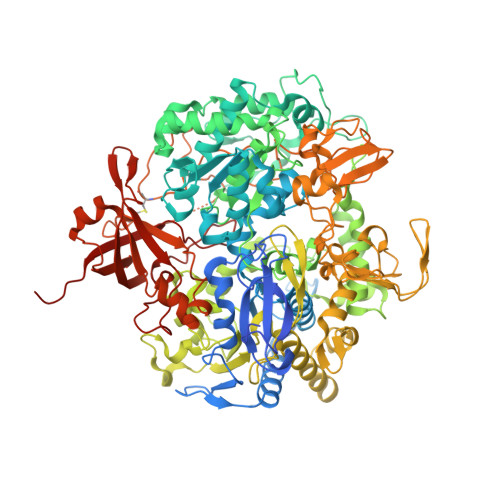 | |
UniProt | |||||
Find proteins for Q72EJ1 (Nitratidesulfovibrio vulgaris (strain ATCC 29579 / DSM 644 / CCUG 34227 / NCIMB 8303 / VKM B-1760 / Hildenborough)) Explore Q72EJ1 Go to UniProtKB: Q72EJ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72EJ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Formate dehydrogenase, beta subunit, putative | 236 | Nitratidesulfovibrio vulgaris str. Hildenborough | Mutation(s): 0 | 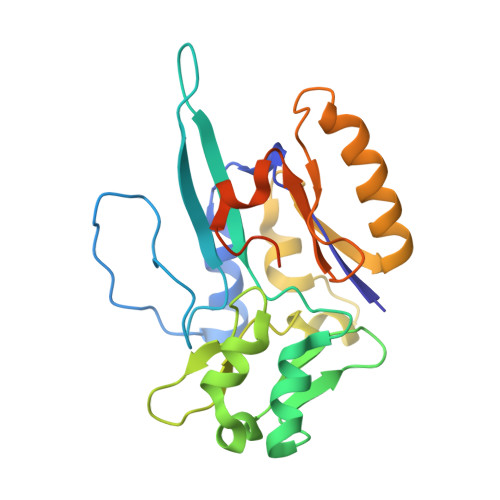 | |
UniProt | |||||
Find proteins for Q72EJ0 (Nitratidesulfovibrio vulgaris (strain ATCC 29579 / DSM 644 / CCUG 34227 / NCIMB 8303 / VKM B-1760 / Hildenborough)) Explore Q72EJ0 Go to UniProtKB: Q72EJ0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72EJ0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MGD (Subject of Investigation/LOI) Query on MGD | C [auth A], D [auth A] | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE C20 H26 N10 O13 P2 S2 VQAGYJCYOLHZDH-ILXWUORBSA-N |  | ||
| SF4 Query on SF4 | E [auth A], U [auth B], V [auth B], W [auth B] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| W (Subject of Investigation/LOI) Query on W | F [auth A] | TUNGSTEN ION W FZFRVZDLZISPFJ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | T [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | H [auth A] I [auth A] J [auth A] K [auth A] L [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| NO3 Query on NO3 | R [auth A], S [auth A] | NITRATE ION N O3 NHNBFGGVMKEFGY-UHFFFAOYSA-N |  | ||
| H2S (Subject of Investigation/LOI) Query on H2S | G [auth A] | HYDROSULFURIC ACID H2 S RWSOTUBLDIXVET-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 64.885 | α = 90 |
| b = 128.56 | β = 90 |
| c = 149.629 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Fundacao para a Ciencia e a Tecnologia | Portugal | PTDC/BBB-EBB/2723/2014 |
| Fundacao para a Ciencia e a Tecnologia | Portugal | UID/Multi/04378/2019 |