Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC).
Ward, R.A., Anderton, M.J., Bethel, P., Breed, J., Cook, C., Davies, E.J., Dobson, A., Dong, Z., Fairley, G., Farrington, P., Feron, L., Flemington, V., Gibbons, F.D., Graham, M.A., Greenwood, R., Hanson, L., Hopcroft, P., Howells, R., Hudson, J., James, M., Jones, C.D., Jones, C.R., Li, Y., Lamont, S., Lewis, R., Lindsay, N., McCabe, J., McGuire, T., Rawlins, P., Roberts, K., Sandin, L., Simpson, I., Swallow, S., Tang, J., Tomkinson, G., Tonge, M., Wang, Z., Zhai, B.(2019) J Med Chem 62: 11004-11018
- PubMed: 31710489
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01295
- Primary Citation of Related Structures:
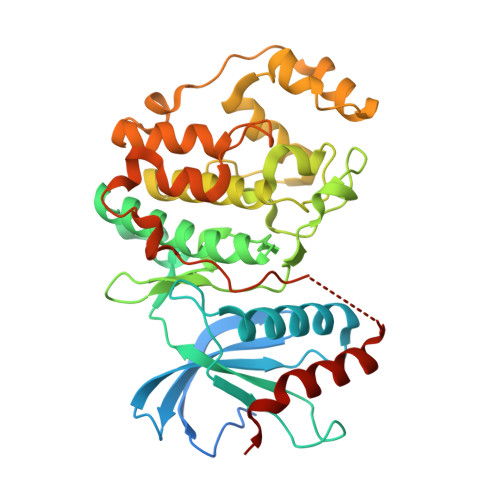
6SLG - PubMed Abstract:
The RAS/MAPK pathway is a major driver of oncogenesis and is dysregulated in approximately 30% of human cancers, primarily by mutations in the BRAF or RAS genes. The extracellular-signal-regulated kinases (ERK1 and ERK2) serve as central nodes within this pathway. The feasibility of targeting the RAS/MAPK pathway has been demonstrated by the clinical responses observed through the use of BRAF and MEK inhibitors in BRAF V600E/K metastatic melanoma; however, resistance frequently develops. Importantly, ERK1/2 inhibition may have clinical utility in overcoming acquired resistance to RAF and MEK inhibitors, where RAS/MAPK pathway reactivation has occurred, such as relapsed BRAF V600E/K melanoma. We describe our structure-based design approach leading to the discovery of AZD0364, a potent and selective inhibitor of ERK1 and ERK2. AZD0364 exhibits high cellular potency (IC 50 = 6 nM) as well as excellent physicochemical and absorption, distribution, metabolism, and excretion (ADME) properties and has demonstrated encouraging antitumor activity in preclinical models.
- Oncology and Discovery Sciences R&D , AstraZeneca , Darwin Building and Hodgkin Building, c/o Darwin Building, 310 Cambridge Science Park, Milton Rd , Cambridge CB4 0WG , U.K.
Organizational Affiliation: