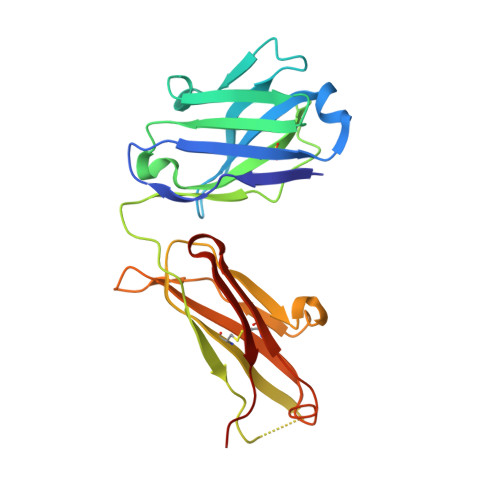
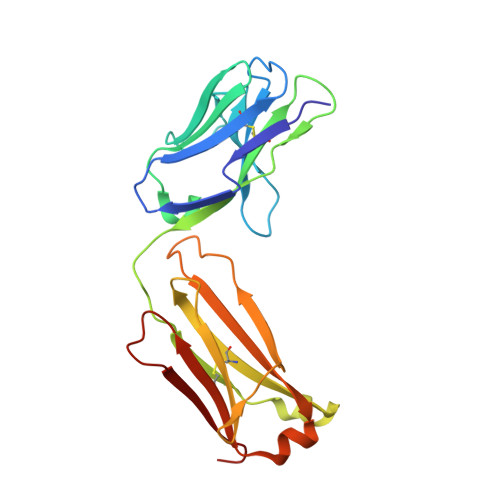
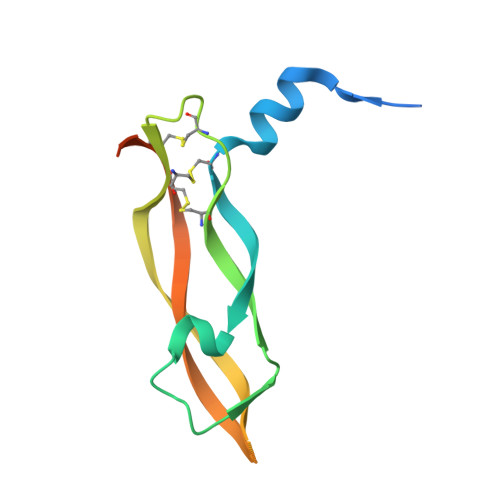
DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Beckmann, R., Jensen, K., Fenn, S., Speck, J., Krause, K., Meier, A., Roth, M., Fauser, S., Kimbung, R., Logan, D.T., Steegmaier, M., Kettenberger, H.(2021) Nat Commun 12: 708-708
- PubMed: 33514724
- DOI: https://doi.org/10.1038/s41467-021-20949-3
- Primary Citation of Related Structures:
6T9D, 6T9E - PubMed Abstract:
We report the development of a platform of dual targeting Fab (DutaFab) molecules, which comprise two spatially separated and independent binding sites within the human antibody CDR loops: the so-called H-side paratope encompassing HCDR1, HCDR3 and LCDR2, and the L-side paratope encompassing LCDR1, LCDR3 and HCDR2. Both paratopes can be independently selected and combined into the desired bispecific DutaFabs in a modular manner. X-ray crystal structures illustrate that DutaFabs are able to bind two target molecules simultaneously at the same Fv region comprising a VH-VL heterodimer. In the present study, this platform is applied to generate DutaFabs specific for VEGFA and PDGF-BB, which show high affinities, physico-chemical stability and solubility, as well as superior efficacy over anti-VEGF monotherapy in vivo. These molecules exemplify the usefulness of DutaFabs as a distinct class of antibody therapeutics, which is currently being evaluated in patients.
- Roche Pharma Research and Early Development, Roche Innovation Center Munich, Roche Diagnostics GmbH, Penzberg, Germany. pred.dutafabs@roche.com.
Organizational Affiliation: