Ultra-large chemical libraries for the discovery of high-affinity peptide binders.
Quartararo, A.J., Gates, Z.P., Somsen, B.A., Hartrampf, N., Ye, X., Shimada, A., Kajihara, Y., Ottmann, C., Pentelute, B.L.(2020) Nat Commun 11: 3183-3183
- PubMed: 32576815
- DOI: https://doi.org/10.1038/s41467-020-16920-3
- Primary Citation of Related Structures:
6TCH - PubMed Abstract:
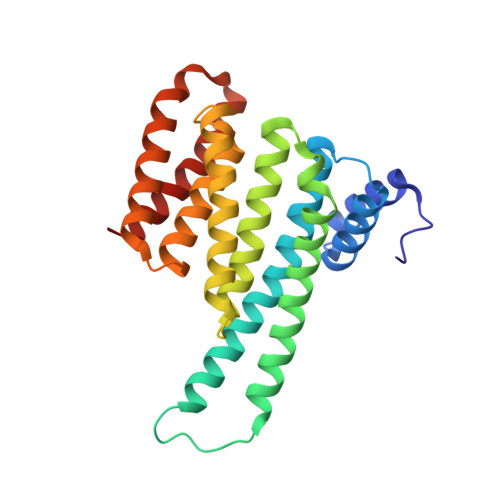
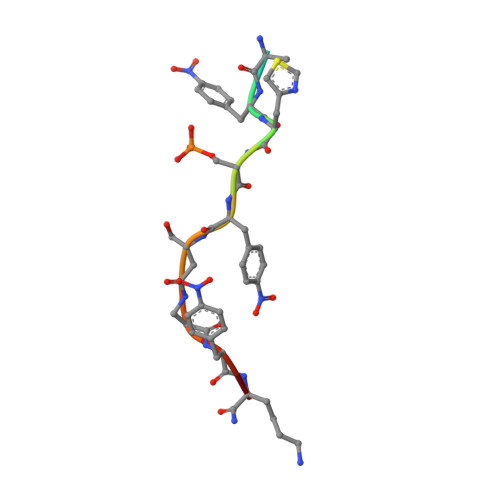
High-diversity genetically-encoded combinatorial libraries (10 8 -10 13 members) are a rich source of peptide-based binding molecules, identified by affinity selection. Synthetic libraries can access broader chemical space, but typically examine only ~ 10 6 compounds by screening. Here we show that in-solution affinity selection can be interfaced with nano-liquid chromatography-tandem mass spectrometry peptide sequencing to identify binders from fully randomized synthetic libraries of 10 8 members-a 100-fold gain in diversity over standard practice. To validate this approach, we show that binders to a monoclonal antibody are identified in proportion to library diversity, as diversity is increased from 10 6 -10 8 . These results are then applied to the discovery of p53-like binders to MDM2, and to a family of 3-19 nM-affinity, α/β-peptide-based binders to 14-3-3. An X-ray structure of one of these binders in complex with 14-3-3σ is determined, illustrating the role of β-amino acids in facilitating a key binding contact.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.
Organizational Affiliation: