Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Bermejo, I.A., Navo, C.D., Castro-Lopez, J., Guerreiro, A., Jimenez-Moreno, E., Sanchez Fernandez, E.M., Garcia-Martin, F., Hinou, H., Nishimura, S.I., Garcia Fernandez, J.M., Mellet, C.O., Avenoza, A., Busto, J.H., Bernardes, G.J.L., Hurtado-Guerrero, R., Peregrina, J.M., Corzana, F.(2020) Chem Sci 11: 3996-4006
- PubMed: 34122869
- DOI: https://doi.org/10.1039/c9sc06334j
- Primary Citation of Related Structures:
6TGG - PubMed Abstract:
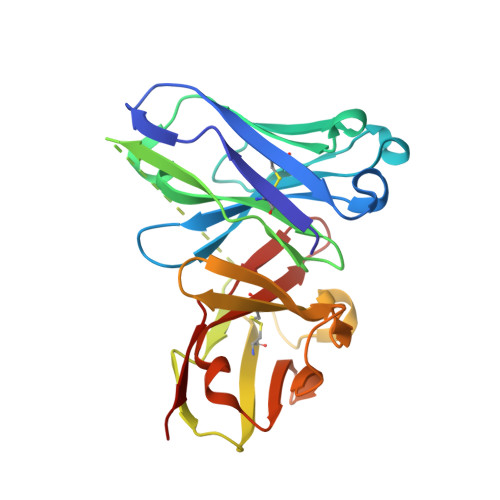
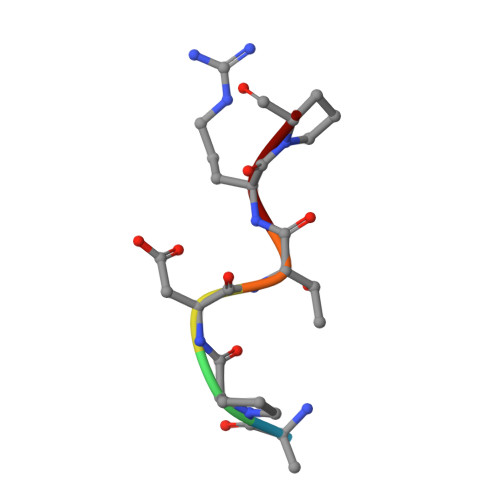
The Tn antigen (GalNAc-α-1- O -Thr/Ser) is a well-known tumor-associated carbohydrate determinant. The use of glycopeptides that incorporate this structure has become a significant and promising niche of research owing to their potential use as anticancer vaccines. Herein, the conformational preferences of a glycopeptide with an unnatural Tn antigen, characterized by a threonine decorated with an sp 2 -iminosugar-type α-GalNAc mimic, have been studied both in solution, by combining NMR spectroscopy and molecular dynamics simulations, and in the solid state bound to an anti-mucin-1 (MUC1) antibody, by X-ray crystallography. The Tn surrogate can mimic the main conformer sampled by the natural antigen in solution and exhibits high affinity towards anti-MUC1 antibodies. Encouraged by these data, a cancer vaccine candidate based on this unnatural glycopeptide and conjugated to the carrier protein Keyhole Limpet Hemocyanin (KLH) has been prepared and tested in mice. Significantly, the experiments in vivo have proved that this vaccine elicits higher levels of specific anti-MUC1 IgG antibodies than the analog that bears the natural Tn antigen and that the elicited antibodies recognize human breast cancer cells with high selectivity. Altogether, we compile evidence to confirm that the presentation of the antigen, both in solution and in the bound state, plays a critical role in the efficacy of the designed cancer vaccines. Moreover, the outcomes derived from this vaccine prove that there is room for exploring further adjustments at the carbohydrate level that could contribute to designing more efficient cancer vaccines.
- Departamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química E-26006 Logroño Spain francisco.corzana@unirioja.es jesusmanuel.peregrina@unirioja.es.
Organizational Affiliation: