Novel non-covalent LSD1 inhibitors endowed with anticancer effects in leukemia and solid tumor cellular models.
Menna, M., Fiorentino, F., Marrocco, B., Lucidi, A., Tomassi, S., Cilli, D., Romanenghi, M., Cassandri, M., Pomella, S., Pezzella, M., Del Bufalo, D., Zeya Ansari, M.S., Tomasevic, N., Mladenovic, M., Viviano, M., Sbardella, G., Rota, R., Trisciuoglio, D., Minucci, S., Mattevi, A., Rotili, D., Mai, A.(2022) Eur J Med Chem 237: 114410-114410
- PubMed: 35525212
- DOI: https://doi.org/10.1016/j.ejmech.2022.114410
- Primary Citation of Related Structures:
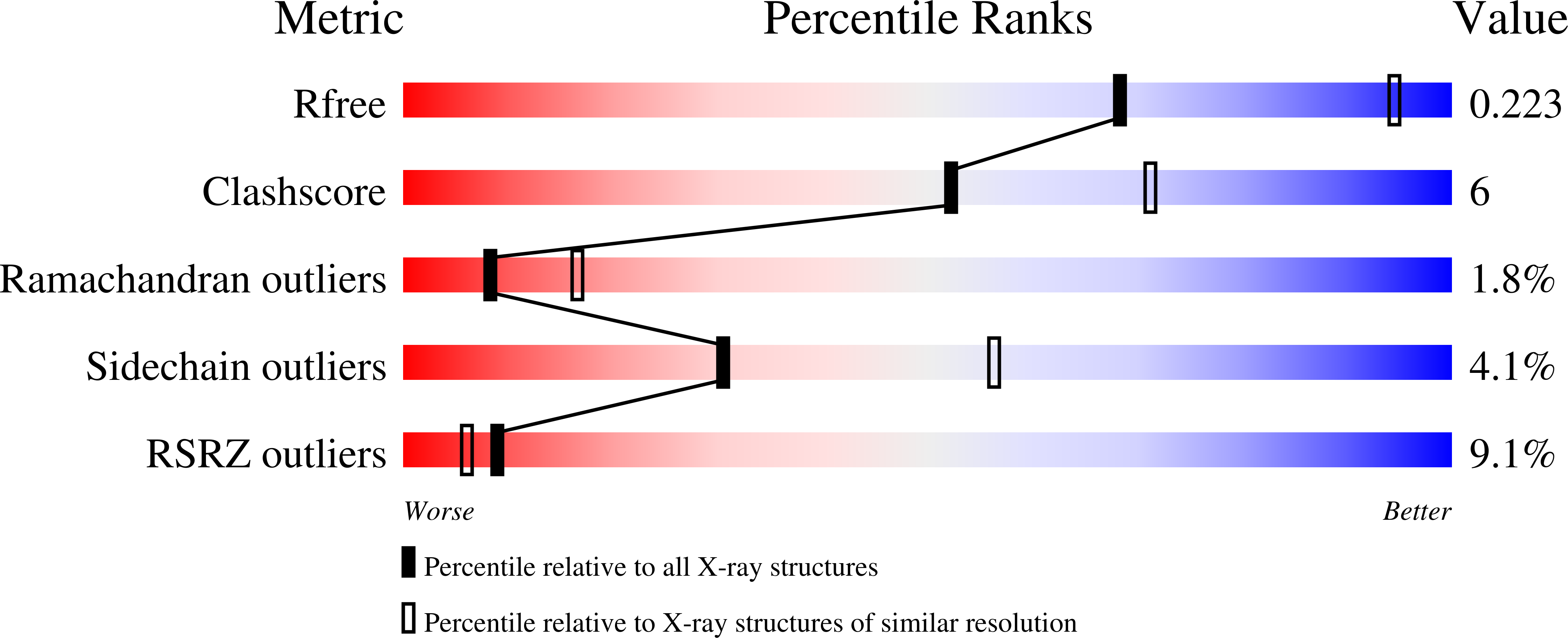
6TUY - PubMed Abstract:
LSD1 is a histone lysine demethylase proposed as therapeutic target in cancer. Chemical modifications applied at C2, C4 and/or C7 positions of the quinazoline core of the previously reported dual LSD1/G9a inhibitor 1 led to a series of non-covalent, highly active, and selective LSD1 inhibitors (2-4 and 6-30) and to the dual LSD1/G9a inhibitor 5 that was more potent than 1 against LSD1. In THP-1 and MV4-11 leukemic cells, the most potent compounds (7, 8, and 29) showed antiproliferative effects at sub-micromolar level without significant toxicity at 1 μM in non-cancer AHH-1 cells. In MV4-11 cells, the new derivatives increased the levels of the LSD1 histone mark H3K4me2 and induced the re-expression of the CD86 gene silenced by LSD1, thereby confirming the inhibition of LSD1 at cellular level. In breast MDA-MB-231 as well as in rhabdomyosarcoma RD and RH30 cells, taken as examples of solid tumors, the same compounds displayed cell growth arrest in the same IC 50 range, highlighting a crucial anticancer role for LSD1 inhibition and suggesting no added value for the simultaneous G9a inhibition in these tumor cell lines.
Organizational Affiliation:
Department of Drug Chemistry and Technologies, Sapienza University of Rome, P. le A. Moro 5, 00185, Rome, Italy.