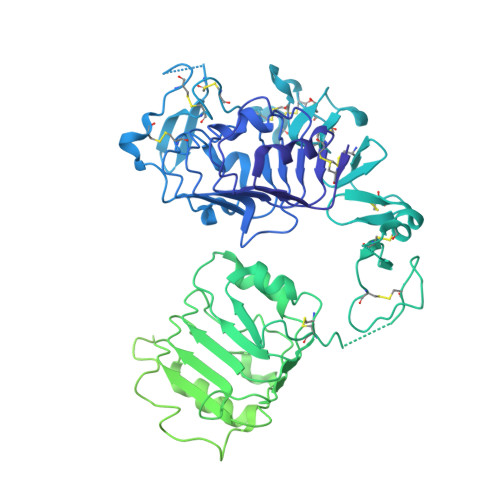
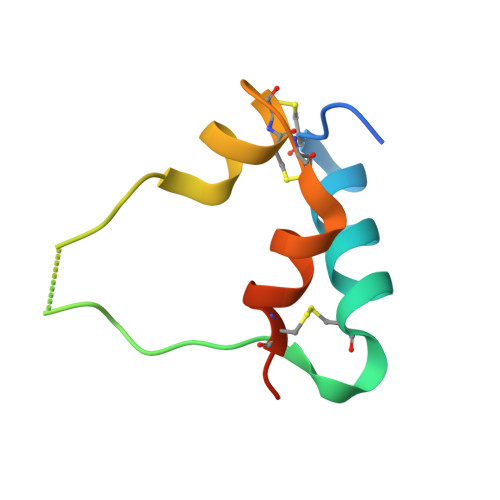
How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Xu, Y., Kirk, N.S., Venugopal, H., Margetts, M.B., Croll, T.I., Sandow, J.J., Webb, A.I., Delaine, C.A., Forbes, B.E., Lawrence, M.C.(2020) Structure 28: 786-798.e6
- PubMed: 32459985
- DOI: https://doi.org/10.1016/j.str.2020.05.002
- Primary Citation of Related Structures:
6VWG, 6VWH, 6VWI, 6VWJ - PubMed Abstract:
Human type 1 insulin-like growth factor receptor (IGF-1R) signals chiefly in response to the binding of insulin-like growth factor I. Relatively little is known about the role of insulin-like growth factor II signaling via IGF-1R, despite the affinity of insulin-like growth factor II for IGF-1R being within an order of magnitude of that of insulin-like growth factor I. Here, we describe the cryoelectron microscopy structure of insulin-like growth factor II bound to a leucine-zipper-stabilized IGF-1R ectodomain, determined in two conformations to a maximum average resolution of 3.2 Å. The two conformations differ in the relative separation of their respective points of membrane entry, and comparison with the structure of insulin-like growth factor I bound to IGF-1R reveals long-suspected differences in the way in which the critical C domain of the respective growth factors interact with IGF-1R.
- Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia; Department of Medical Biology, Faculty of Medicine, Dentistry and Health Sciences, University of Melbourne, Parkville, VIC 3050, Australia.
Organizational Affiliation: