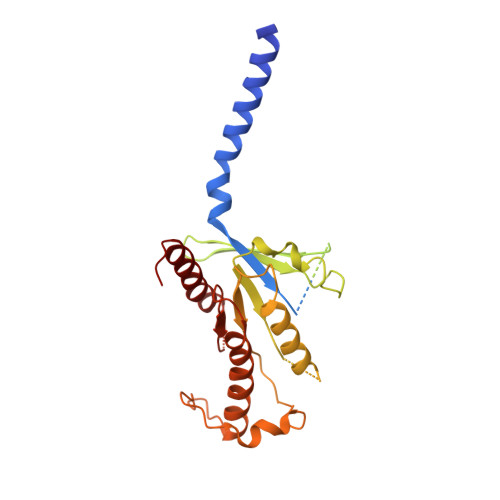
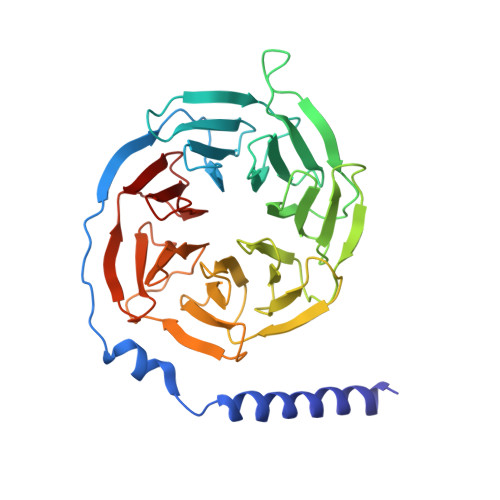
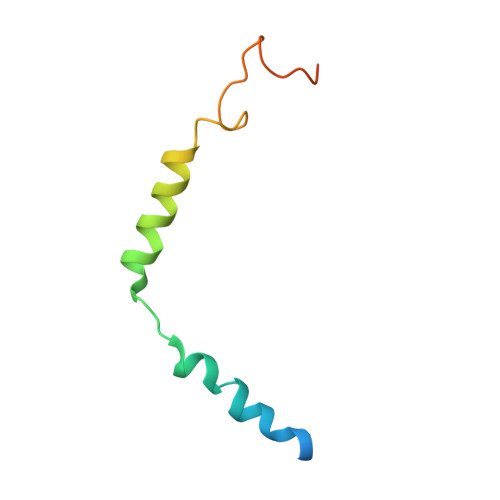
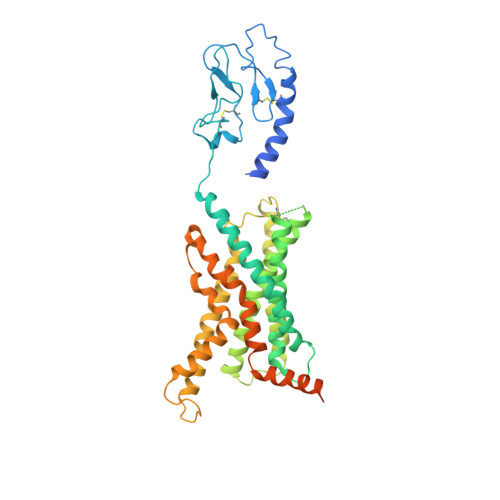
Structure and dynamics of the active Gs-coupled human secretin receptor.
Dong, M., Deganutti, G., Piper, S.J., Liang, Y.L., Khoshouei, M., Belousoff, M.J., Harikumar, K.G., Reynolds, C.A., Glukhova, A., Furness, S.G.B., Christopoulos, A., Danev, R., Wootten, D., Sexton, P.M., Miller, L.J.(2020) Nat Commun 11: 4137-4137
- PubMed: 32811827
- DOI: https://doi.org/10.1038/s41467-020-17791-4
- Primary Citation of Related Structures:
6WI9, 6WZG - PubMed Abstract:
The class B secretin GPCR (SecR) has broad physiological effects, with target potential for treatment of metabolic and cardiovascular disease. Molecular understanding of SecR binding and activation is important for its therapeutic exploitation. We combined cryo-electron microscopy, molecular dynamics, and biochemical cross-linking to determine a 2.3 Å structure, and interrogate dynamics, of secretin bound to the SecR:Gs complex. SecR exhibited a unique organization of its extracellular domain (ECD) relative to its 7-transmembrane (TM) core, forming more extended interactions than other family members. Numerous polar interactions formed between secretin and the receptor extracellular loops (ECLs) and TM helices. Cysteine-cross-linking, cryo-electron microscopy multivariate analysis and molecular dynamics simulations revealed that interactions between peptide and receptor were dynamic, and suggested a model for initial peptide engagement where early interactions between the far N-terminus of the peptide and SecR ECL2 likely occur following initial binding of the peptide C-terminus to the ECD.
- Department of Molecular Pharmacology and Experimental Therapeutics, Mayo Clinic, Scottsdale, AZ, 85259, USA.
Organizational Affiliation: