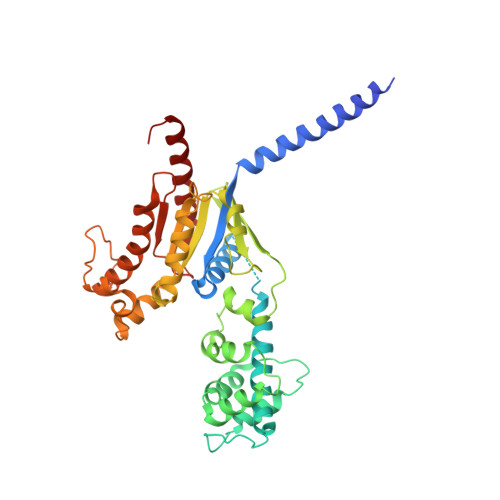
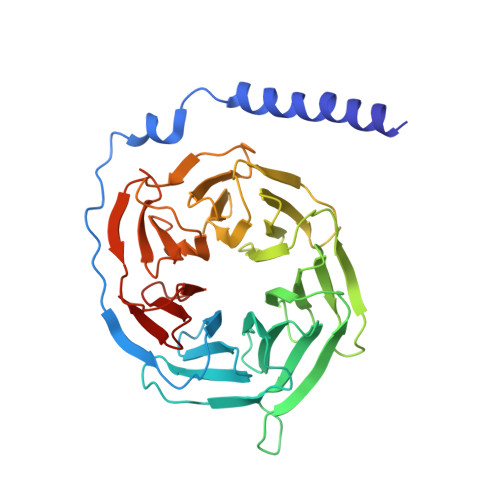
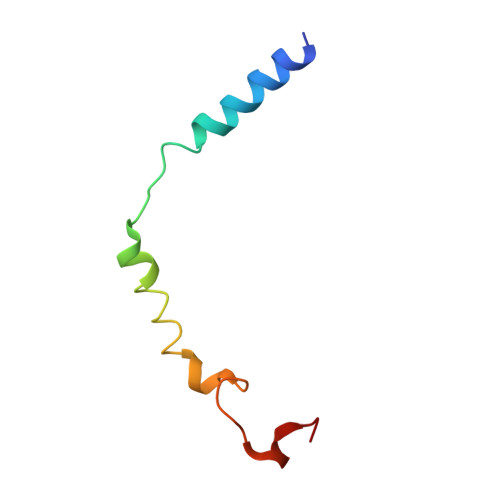
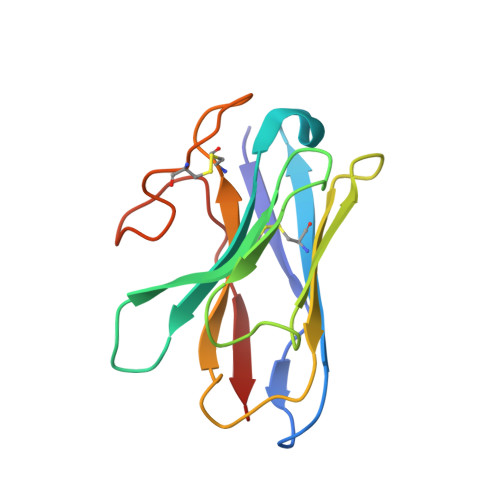
Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Zhang, X., Belousoff, M.J., Zhao, P., Kooistra, A.J., Truong, T.T., Ang, S.Y., Underwood, C.R., Egebjerg, T., Senel, P., Stewart, G.D., Liang, Y.L., Glukhova, A., Venugopal, H., Christopoulos, A., Furness, S.G.B., Miller, L.J., Reedtz-Runge, S., Langmead, C.J., Gloriam, D.E., Danev, R., Sexton, P.M., Wootten, D.(2020) Mol Cell 80: 485
- PubMed: 33027691
- DOI: https://doi.org/10.1016/j.molcel.2020.09.020
- Primary Citation of Related Structures:
6X18, 6X19, 6X1A - PubMed Abstract:
Peptide drugs targeting class B1 G-protein-coupled receptors (GPCRs) can treat multiple diseases; however, there remains substantial interest in the development of orally delivered non-peptide drugs. Here, we reveal unexpected overlap between signaling and regulation of the glucagon-like peptide-1 (GLP-1) receptor by the non-peptide agonist PF 06882961 and GLP-1 that was not observed for another compound, CHU-128. Compounds from these patent series, including PF 06882961, are currently in clinical trials for treatment of type 2 diabetes. High-resolution cryoelectron microscopy (cryo-EM) structures reveal that the binding sites for PF 06882961 and GLP-1 substantially overlap, whereas CHU-128 adopts a unique binding mode with a more open receptor conformation at the extracellular face. Structural differences involving extensive water-mediated hydrogen bond networks could be correlated to functional data to understand how PF 06882961, but not CHU-128, can closely mimic the pharmacological properties of GLP-1. These findings will facilitate rational structure-based discovery of non-peptide agonists targeting class B GPCRs.
- Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC 3052, Australia.
Organizational Affiliation: