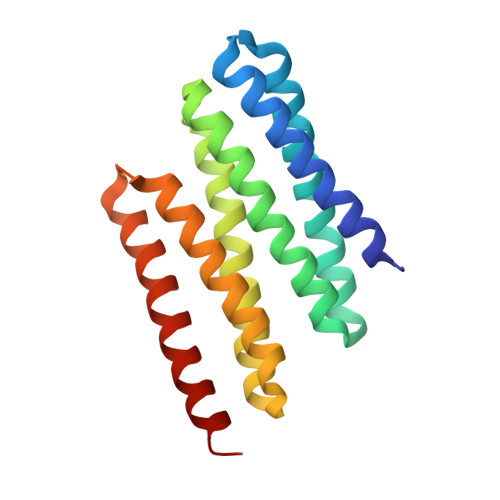
Design of functionalised circular tandem repeat proteins with longer repeat topologies and enhanced subunit contact surfaces.
Hallinan, J.P., Doyle, L.A., Shen, B.W., Gewe, M.M., Takushi, B., Kennedy, M.A., Friend, D., Roberts, J.M., Bradley, P., Stoddard, B.L.(2021) Commun Biol 4: 1240-1240
- PubMed: 34716407
- DOI: https://doi.org/10.1038/s42003-021-02766-y
- Primary Citation of Related Structures:
6XR1, 6XR2, 7RDR - PubMed Abstract:
Circular tandem repeat proteins ('cTRPs') are de novo designed protein scaffolds (in this and prior studies, based on antiparallel two-helix bundles) that contain repeated protein sequences and structural motifs and form closed circular structures. They can display significant stability and solubility, a wide range of sizes, and are useful as protein display particles for biotechnology applications. However, cTRPs also demonstrate inefficient self-assembly from smaller subunits. In this study, we describe a new generation of cTRPs, with longer repeats and increased interaction surfaces, which enhanced the self-assembly of two significantly different sizes of homotrimeric constructs. Finally, we demonstrated functionalization of these constructs with (1) a hexameric array of peptide-binding SH2 domains, and (2) a trimeric array of anti-SARS CoV-2 VHH domains. The latter proved capable of sub-nanomolar binding affinities towards the viral receptor binding domain and potent viral neutralization function.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave. North, Seattle, WA, 98109, USA.
Organizational Affiliation: