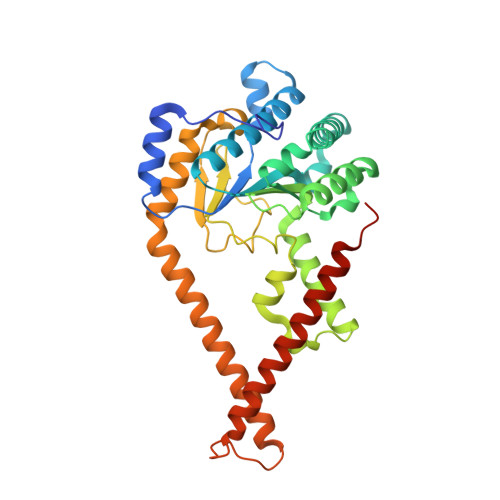
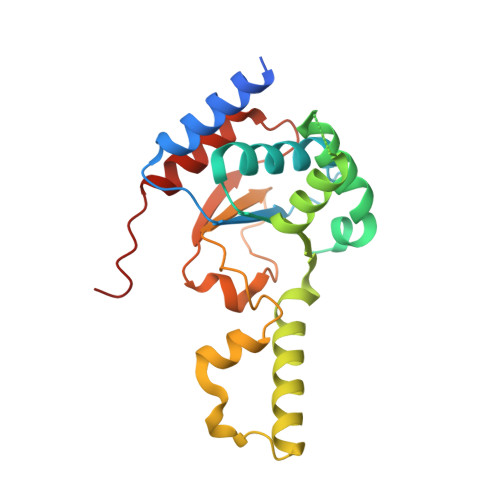
Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase complex.
Bar-El, M.L., Vankova, P., Yeheskel, A., Simhaev, L., Engel, H., Man, P., Haitin, Y., Giladi, M.(2020) Nat Commun 11: 5273-5273
- PubMed: 33077723
- DOI: https://doi.org/10.1038/s41467-020-18970-z
- Primary Citation of Related Structures:
6Z1N - PubMed Abstract:
The human cis-prenyltransferase (hcis-PT) is an enzymatic complex essential for protein N-glycosylation. Synthesizing the precursor of the glycosyl carrier dolichol-phosphate, mutations in hcis-PT cause severe human diseases. Here, we reveal that hcis-PT exhibits a heterotetrameric assembly in solution, consisting of two catalytic dehydrodolichyl diphosphate synthase (DHDDS) and inactive Nogo-B receptor (NgBR) heterodimers. Importantly, the 2.3 Å crystal structure reveals that the tetramer assembles via the DHDDS C-termini as a dimer-of-heterodimers. Moreover, the distal C-terminus of NgBR transverses across the interface with DHDDS, directly participating in active-site formation and the functional coupling between the subunits. Finally, we explored the functional consequences of disease mutations clustered around the active-site, and in combination with molecular dynamics simulations, we propose a mechanism for hcis-PT dysfunction in retinitis pigmentosa. Together, our structure of the hcis-PT complex unveils the dolichol synthesis mechanism and its perturbation in disease.
- Department of Physiology and Pharmacology, Sackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv, 6997801, Israel.
Organizational Affiliation: