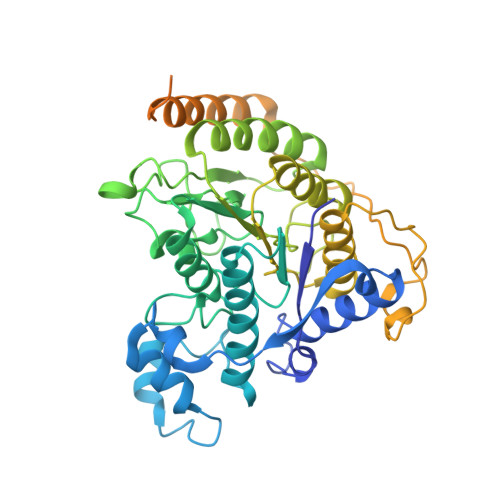
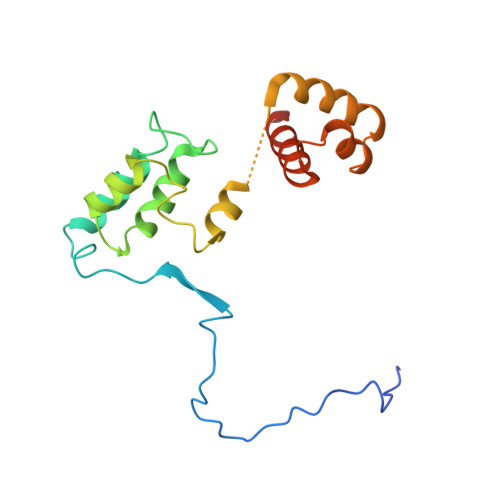
The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Turnbull, R.E., Fairall, L., Saleh, A., Kelsall, E., Morris, K.L., Ragan, T.J., Savva, C.G., Chandru, A., Millard, C.J., Makarova, O.V., Smith, C.J., Roseman, A.M., Fry, A.M., Cowley, S.M., Schwabe, J.W.R.(2020) Nat Commun 11: 3252-3252
- PubMed: 32591534
- DOI: https://doi.org/10.1038/s41467-020-17078-8
- Primary Citation of Related Structures:
6Z2J, 6Z2K - PubMed Abstract:
MiDAC is one of seven distinct, large multi-protein complexes that recruit class I histone deacetylases to the genome to regulate gene expression. Despite implications of involvement in cell cycle regulation and in several cancers, surprisingly little is known about the function or structure of MiDAC. Here we show that MiDAC is important for chromosome alignment during mitosis in cancer cell lines. Mice lacking the MiDAC proteins, DNTTIP1 or MIDEAS, die with identical phenotypes during late embryogenesis due to perturbations in gene expression that result in heart malformation and haematopoietic failure. This suggests that MiDAC has an essential and unique function that cannot be compensated by other HDAC complexes. Consistent with this, the cryoEM structure of MiDAC reveals a unique and distinctive mode of assembly. Four copies of HDAC1 are positioned at the periphery with outward-facing active sites suggesting that the complex may target multiple nucleosomes implying a processive deacetylase function.
- Leicester Institute of Structural and Chemical Biology, University of Leicester, Leicester, LE1 7RH, UK.
Organizational Affiliation: