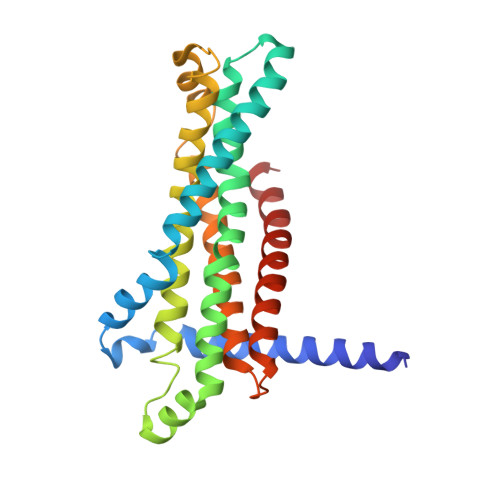
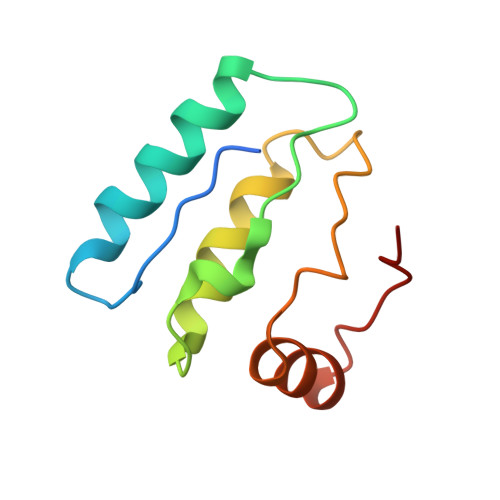
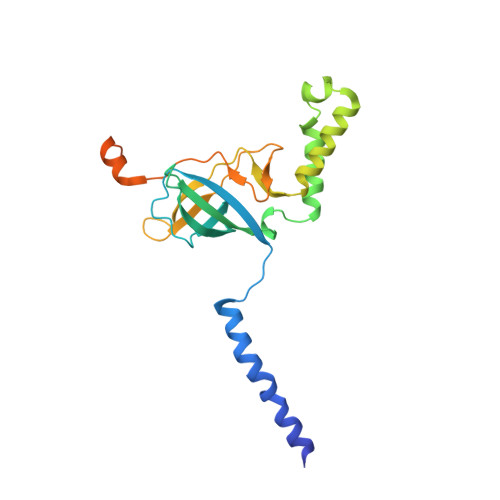
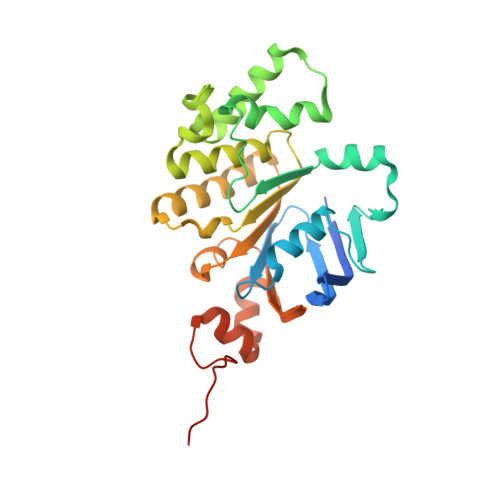
Structure and lipid dynamics in the maintenance of lipid asymmetry inner membrane complex of A. baumannii.
Mann, D., Fan, J., Somboon, K., Farrell, D.P., Muenks, A., Tzokov, S.B., DiMaio, F., Khalid, S., Miller, S.I., Bergeron, J.R.C.(2021) Commun Biol 4: 817-817
- PubMed: 34188171
- DOI: https://doi.org/10.1038/s42003-021-02318-4
- Primary Citation of Related Structures:
6Z5U - PubMed Abstract:
Multi-resistant bacteria are a major threat in modern medicine. The gram-negative coccobacillus Acinetobacter baumannii currently leads the WHO list of pathogens in critical need for new therapeutic development. The maintenance of lipid asymmetry (MLA) protein complex is one of the core machineries that transport lipids from/to the outer membrane in gram-negative bacteria. It also contributes to broad-range antibiotic resistance in several pathogens, most prominently in A. baumannii. Nonetheless, the molecular details of its role in lipid transport has remained largely elusive. Here, we report the cryo-EM maps of the core MLA complex, MlaBDEF, from the pathogen A. baumannii, in the apo-, ATP- and ADP-bound states, revealing multiple lipid binding sites in the cytosolic and periplasmic side of the complex. Molecular dynamics simulations suggest their potential trajectory across the membrane. Collectively with the recently-reported structures of the E. coli orthologue, this data also allows us to propose a molecular mechanism of lipid transport by the MLA system.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, The University of Sheffield, Sheffield, UK.