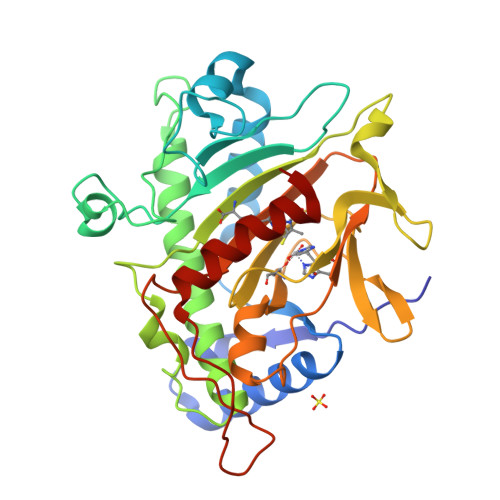
Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
Rabe, P., Kamps, J.J.A.G., Sutherlin, K., Pharm, C., McDonough, M.A., Leissing, T.M., Aller, P., Butryn, A., Linyard, J., Lang, P., Brem, J., Fuller, F.D., Batyuk, A., Hunter, M.S., Pettinati, I., Clifton, I.J., Alonso-Mori, R., Gul, S., Young, I., Kim, I., Bhowmick, A., ORiordan, L., Brewster, A.S., Claridge, T.D.W., Sauter, N.K., Yachandra, V., Yano, J., Kern, J.F., Orville, A.M., Schofield, C.J.To be published.