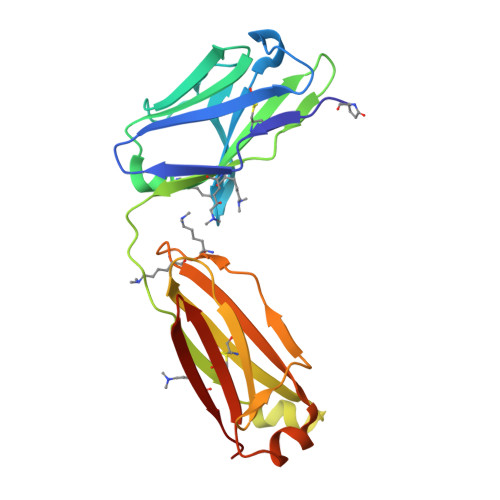
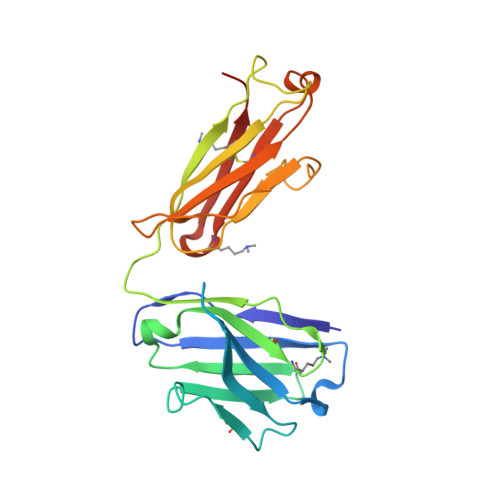
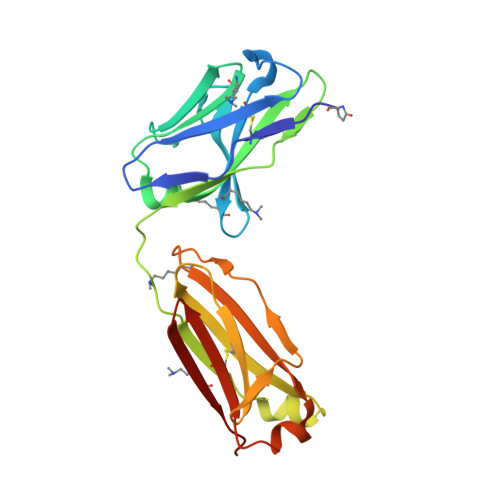
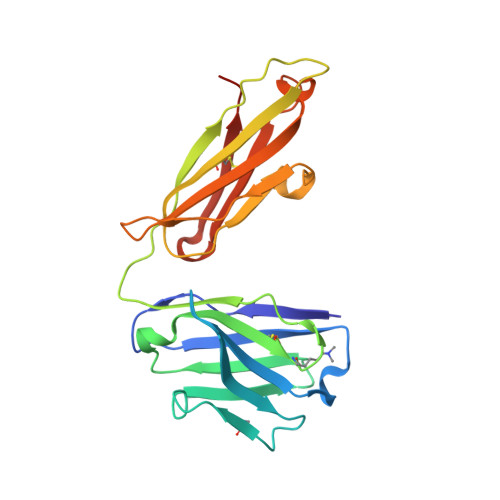
Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Tan, K., Zhou, M., Ahrendt, A.J., Duke, N.E.C., Tabaja, N., Ball, W.J., Kirley, T.L., Norman, A.B., Joachimiak, A., Schiffer, M., Wilton, R., Pokkuluri, P.R.(2019) Acta Crystallogr F Struct Biol Commun 75: 697-706
- PubMed: 31702583
- DOI: https://doi.org/10.1107/S2053230X19013608
- Primary Citation of Related Structures:
6NEX, 6NFN - PubMed Abstract:
A high-affinity anti-cocaine monoclonal antibody, designated h2E2, is entering phase 1 clinical trials for cocaine abuse therapy. To gain insight into the molecular details of its structure that are important for binding cocaine and cocaine metabolites, the Fab fragment was generated and crystallized with and without ligand. Structures of the unliganded Fab and the Fab fragment bound to benzoylecgonine were determined, and were compared with each other and with other crystallized anti-cocaine antibodies. The affinity of the h2E2 antibody for cocaine is 4 nM, while that of the cocaine metabolite benzoylecgonine is 20 nM. Both are higher than the reported affinity for cocaine of the two previously crystallized anti-cocaine antibodies. Consistent with cocaine fluorescent quenching binding studies for the h2E2 mAb, four aromatic residues in the CDR regions of the Fab (TyrL32, TyrL96, TrpL91 and TrpH33) were found to be involved in ligand binding. The aromatic side chains surround and trap the tropane moiety of the ligand in the complex structure, forming significant van der Waals interactions which may account for the higher affinity observed for the h2E2 antibody. A water molecule mediates hydrogen bonding between the antibody and the carbonyl group of the benzoyl ester. The affinity of binding to h2E2 of benzoylecgonine differs only by a factor of five compared with that of cocaine; therefore, it is suggested that h2E2 would bind cocaine in the same way as observed in the Fab-benzoylecgonine complex, with minor rearrangements of some hypervariable segments of the antibody.
- Structural Biology Center, X-ray Science Division, Argonne National Laboratory, Lemont, IL 60439, USA.
Organizational Affiliation: