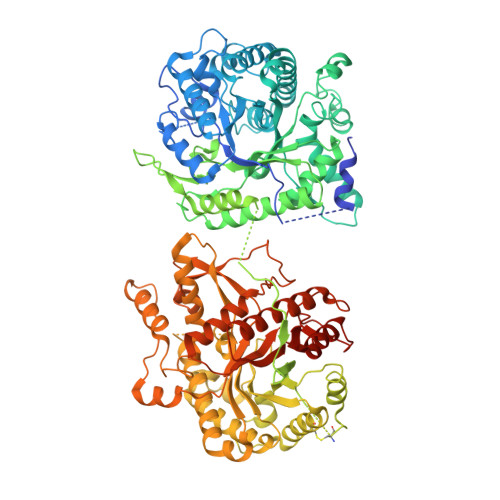
Structures of ligand-occupied beta-Klotho complexes reveal a molecular mechanism underlying endocrine FGF specificity and activity.
Kuzina, E.S., Ung, P.M., Mohanty, J., Tome, F., Choi, J., Pardon, E., Steyaert, J., Lax, I., Schlessinger, A., Schlessinger, J., Lee, S.(2019) Proc Natl Acad Sci U S A 116: 7819-7824
- PubMed: 30944224
- DOI: https://doi.org/10.1073/pnas.1822055116
- Primary Citation of Related Structures:
6NFJ - PubMed Abstract:
The three members of the endocrine fibroblast growth factor (FGF) family designated FGF19, FGF21, and FGF23 mediate their pleiotropic cellular effects by binding to and activating binary complexes composed of an FGF receptor (FGFR) bound to either α-Klotho or β-Klotho receptors. Structural analyses of ligand-occupied Klotho extracellular domains have provided important insights concerning mechanisms underlying the binding specificities of FGF21 and FGF23 to β-Klotho or α-Klotho, respectively. They have also demonstrated that Klotho proteins function as primary high-affinity receptors while FGFRs function as the catalytic subunits that mediate intracellular signaling. Here we describe the crystal structure the C-terminal tail of FGF19 (FGF19 CT ) bound to sKLB and demonstrate that FGF19 CT and FGF21 CT bind to the same binding site on sKLB, via a multiturn D-P motif to site 1 and via a S-P-S motif to the pseudoglycoside hydrolase region (site 2). Binding affinities to sKLB and cellular stimulatory activities of FGF19 CT, FGF21 CT , and a variety of chimeric mutants to cells expressing β-Klotho together with FGFR1c or FGFR4 were also analyzed. These experiments as well as detailed comparison of the structures of free and ligand-occupied sKLB to the structure of ligand-occupied sKLA reveal a general mechanism for recognition of endocrine FGFs by Klotho proteins and regulatory interactions with FGFRs that control their pleiotropic cellular responses.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520.
Organizational Affiliation: