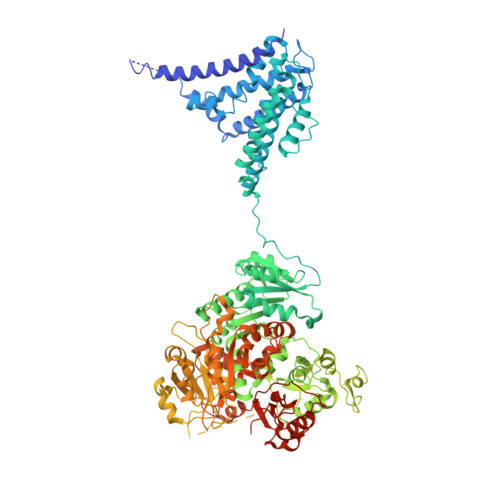
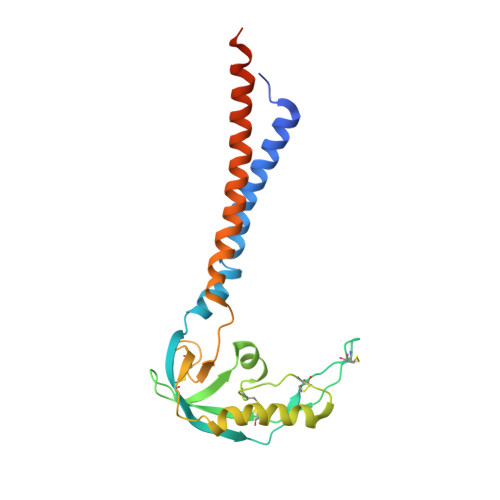
Molecular structures of the human Slo1 K + channel in complex with beta 4.
Tao, X., MacKinnon, R.(2019) Elife 8
- PubMed: 31815672
- DOI: https://doi.org/10.7554/eLife.51409
- Primary Citation of Related Structures:
6V22, 6V35, 6V38, 6V3G - PubMed Abstract:
Slo1 is a Ca 2+ - and voltage-activated K + channel that underlies skeletal and smooth muscle contraction, audition, hormone secretion and neurotransmitter release. In mammals, Slo1 is regulated by auxiliary proteins that confer tissue-specific gating and pharmacological properties. This study presents cryo-EM structures of Slo1 in complex with the auxiliary protein, β4. Four β4, each containing two transmembrane helices, encircle Slo1, contacting it through helical interactions inside the membrane. On the extracellular side, β4 forms a tetrameric crown over the pore. Structures with high and low Ca 2+ concentrations show that identical gating conformations occur in the absence and presence of β4, implying that β4 serves to modulate the relative stabilities of 'pre-existing' conformations rather than creating new ones. The effects of β4 on scorpion toxin inhibition kinetics are explained by the crown, which constrains access but does not prevent binding.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, Howard Hughes Medical Institute, New York, United States.
Organizational Affiliation: