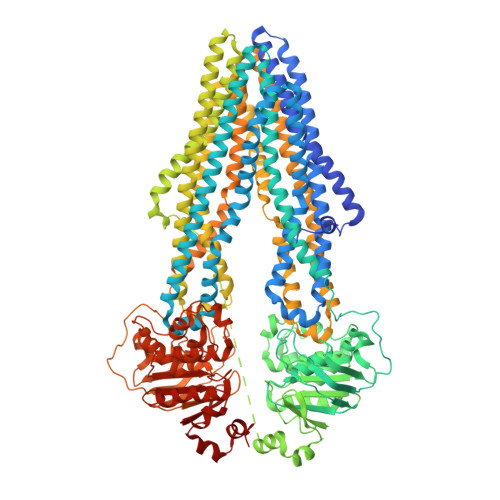
Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1.
Nosol, K., Romane, K., Irobalieva, R.N., Alam, A., Kowal, J., Fujita, N., Locher, K.P.(2020) Proc Natl Acad Sci U S A 117: 26245-26253
- PubMed: 33020312
- DOI: https://doi.org/10.1073/pnas.2010264117
- Primary Citation of Related Structures:
7A65, 7A69, 7A6C, 7A6E, 7A6F - PubMed Abstract:
ABCB1 detoxifies cells by exporting diverse xenobiotic compounds, thereby limiting drug disposition and contributing to multidrug resistance in cancer cells. Multiple small-molecule inhibitors and inhibitory antibodies have been developed for therapeutic applications, but the structural basis of their activity is insufficiently understood. We determined cryo-EM structures of nanodisc-reconstituted, human ABCB1 in complex with the Fab fragment of the inhibitory, monoclonal antibody MRK16 and bound to a substrate (the antitumor drug vincristine) or to the potent inhibitors elacridar, tariquidar, or zosuquidar. We found that inhibitors bound in pairs, with one molecule lodged in the central drug-binding pocket and a second extending into a phenylalanine-rich cavity that we termed the "access tunnel." This finding explains how inhibitors can act as substrates at low concentration, but interfere with the early steps of the peristaltic extrusion mechanism at higher concentration. Our structural data will also help the development of more potent and selective ABCB1 inhibitors.
- Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.
Organizational Affiliation: