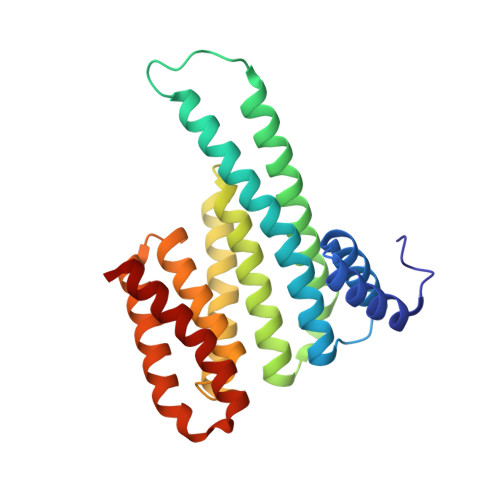
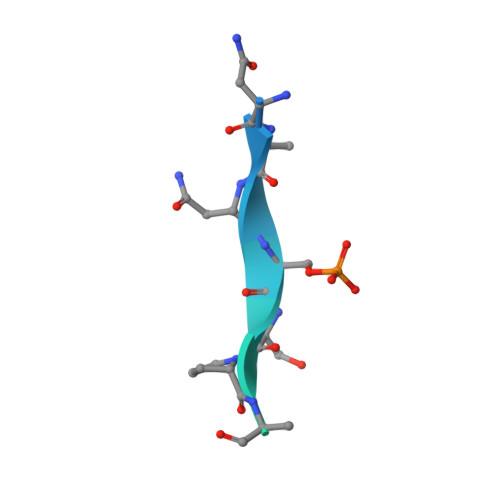
MMP activation-associated aminopeptidase N reveals a bivalent 14-3-3 binding motif.
Kiehstaller, S., Ottmann, C., Hennig, S.(2020) J Biological Chem 295: 18266-18275
- PubMed: 33109610
- DOI: https://doi.org/10.1074/jbc.RA120.014708
- Primary Citation of Related Structures:
6XWD, 7AEW - PubMed Abstract:
Aminopeptidase N (APN, CD13) is a transmembrane ectopeptidase involved in many crucial cellular functions. Besides its role as a peptidase, APN also mediates signal transduction and is involved in the activation of matrix metalloproteinases (MMPs). MMPs function in tissue remodeling within the extracellular space and are therefore involved in many human diseases, such as fibrosis, rheumatoid arthritis, tumor angiogenesis, and metastasis, as well as viral infections. However, the exact mechanism that leads to APN-driven MMP activation is unclear. It was previously shown that extracellular 14-3-3 adapter proteins bind to APN and thereby induce the transcription of MMPs. As a first step, we sought to identify potential 14-3-3-binding sites in the APN sequence. We constructed a set of phosphorylated peptides derived from APN to probe for interactions. We identified and characterized a canonical 14-3-3-binding site ( site 1 ) within the flexible, structurally unresolved N-terminal APN region using direct binding fluorescence polarization assays and thermodynamic analysis. In addition, we identified a secondary, noncanonical binding site ( site 2 ), which enhances the binding affinity in combination with site 1 by many orders of magnitude. Finally, we solved crystal structures of 14-3-3σ bound to mono- and bis-phosphorylated APN-derived peptides, which revealed atomic details of the binding mode of mono- and bivalent 14-3-3 interactions. Therefore, our findings shed some light on the first steps of APN-mediated MMP activation and open the field for further investigation of this important signaling pathway.
- Department of Chemistry and Pharmaceutical Sciences, VU University Amsterdam, Amsterdam, Netherlands; Amsterdam Institute of Molecular and Life Sciences (AIMMS), VU University Amsterdam, Amsterdam, Netherlands.
Organizational Affiliation: