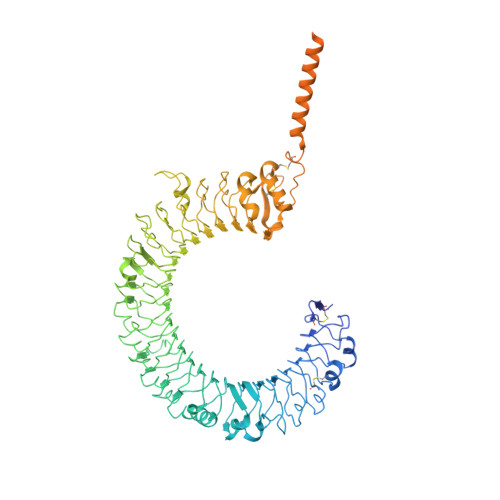
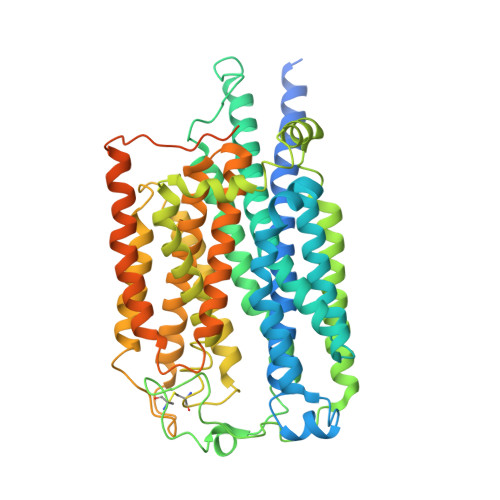
Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Ishida, H., Asami, J., Zhang, Z., Nishizawa, T., Shigematsu, H., Ohto, U., Shimizu, T.(2021) Nat Struct Mol Biol 28: 173-180
- PubMed: 33432245
- DOI: https://doi.org/10.1038/s41594-020-00542-w
- Primary Citation of Related Structures:
7C76, 7C77, 7CYN - PubMed Abstract:
Nucleic acid-sensing Toll-like receptors (TLRs) play a pivotal role in innate immunity by recognizing foreign DNA and RNA. Compartmentalization of these TLRs in the endosome limits their activation by self-derived nucleic acids and reduces the possibility of autoimmune reactions. Although chaperone Unc-93 homolog B1, TLR signaling regulator (UNC93B1) is indispensable for the trafficking of TLRs from the endoplasmic reticulum to the endosome, mechanisms of UNC93B1-mediated TLR regulation remain largely unknown. Here, we report two cryo-EM structures of human and mouse TLR3-UNC93B1 complexes and a human TLR7-UNC93B1 complex. UNC93B1 exhibits structural similarity to the major facilitator superfamily transporters. Both TLRs interact with the UNC93B1 amino-terminal six-helix bundle through their transmembrane and luminal juxtamembrane regions, but the complexes of TLR3 and TLR7 with UNC93B1 differ in their oligomerization state. The structural information provided here should aid in designing compounds to combat autoimmune diseases.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: