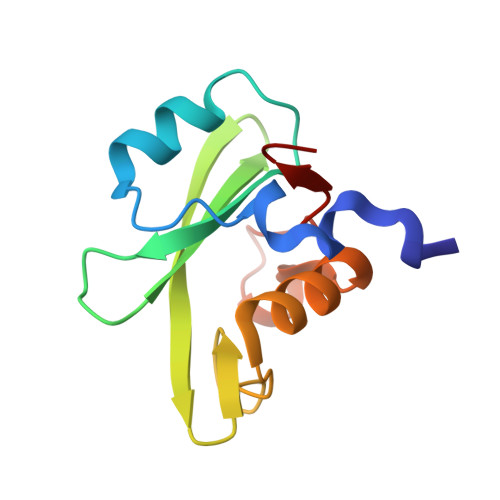
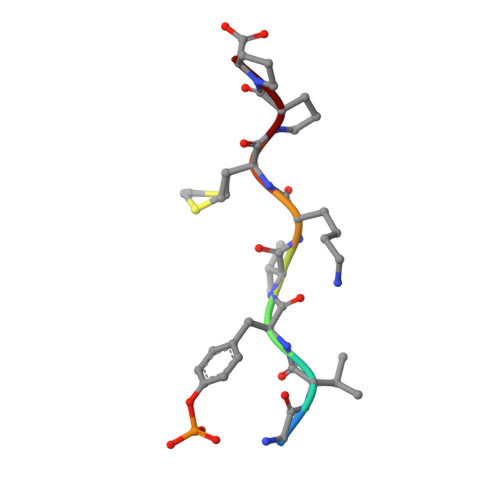
Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains.
Iiyama, M., Numoto, N., Ogawa, S., Kuroda, M., Morii, H., Abe, R., Ito, N., Oda, M.(2021) Mol Immunol 131: 51-59
- PubMed: 33386150
- DOI: https://doi.org/10.1016/j.molimm.2020.12.002
- Primary Citation of Related Structures:
7CIO - PubMed Abstract:
During T-cell regulation, T-cell receptors and CD28 lead to signaling activation, while T-lymphocyte antigen 4 (CTLA-4) is known to lead to downregulation, similar to programmed cell death-1 (PD-1). In the cytoplasmic tails of CD28 and CTLA-4, phosphoinositide 3-kinase (PI3K) binds to the consensus sequence including phosphotyrosine via SH2 domains, N- and C-terminal SH2 domains (nSH2 and cSH2), of its regulatory subunit, p85. In this study, we determined the crystal structure of a CTLA-4-derived phosphopeptide in complex with a Cys-substituted mutant of cSH2, C656S/C659V/C670L, at a 1.1 Å resolution. Phosphotyrosine of the bound peptide is tightly accommodated by the residues Arg631, Arg649, Ser651, and Ser652, similar to the cSH2 wild-type recognition mode of CD28, as reported previously. Upon the Cys mutation, the cSH2 thermal stability increased while the CTLA-4 binding affinity slightly changed. The binding experiments also showed that the binding affinity of CTLA-4 by cSH2 was approximately two orders of magnitude lower than that of CD28. Similar to CD28 binding, the CTLA-4 binding affinity of nSH2 was lower than that of cSH2. The complex structure of nSH2 and CTLA-4 was modeled, and compared with the crystal structure of cSH2 mutant and CTLA-4. The difference in the binding affinity between CD28 and CTLA-4, along with the difference between nSH2 and cSH2, could be explained by the 3D structures, which would be closely correlated with the respective T-cell signaling.
- Graduate School of Life and Environmental Sciences, Kyoto Prefectural University, 1-5 Hangi-cho, Shimogamo, Sakyo-ku, Kyoto 606-8522, Japan.
Organizational Affiliation: