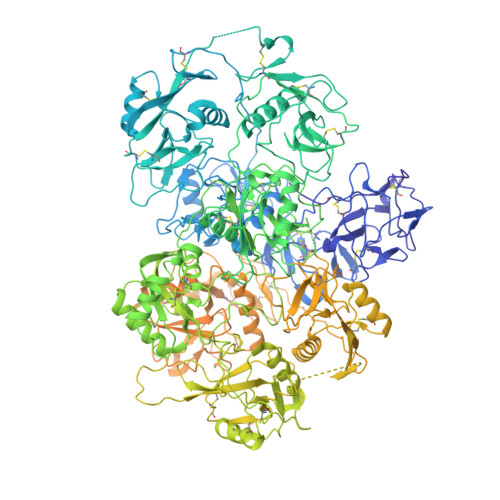
The cryo-EM structure of the endocytic receptor DEC-205.
Gully, B.S., Venugopal, H., Fulcher, A.J., Fu, Z., Li, J., Deuss, F.A., Llerena, C., Heath, W.R., Lahoud, M.H., Caminschi, I., Rossjohn, J., Berry, R.(2020) J Biological Chem 296: 100127-100127
- PubMed: 33257321
- DOI: https://doi.org/10.1074/jbc.RA120.016451
- Primary Citation of Related Structures:
7JPT, 7JPU - PubMed Abstract:
DEC-205 (CD205), a member of the macrophage mannose receptor protein family, is the prototypic endocytic receptor of dendritic cells, whose ligands include phosphorothioated cytosine-guanosine oligonucleotides, a motif often seen in bacterial or viral DNA. However, despite growing biological and clinical significance, little is known about the structural arrangement of this receptor or any of its family members. Here, we describe the 3.2 Å cryo-EM structure of human DEC-205, thereby illuminating the structure of the mannose receptor protein family. The DEC-205 monomer forms a compact structure comprising two intercalated rings of C-type lectin-like domains, where the N-terminal cysteine-rich and fibronectin domains reside at the central intersection. We establish a pH-dependent oligomerization pathway forming tetrameric DEC-205 using solution-based techniques and ultimately solved the 4.9 Å cryo-EM structure of the DEC-205 tetramer to identify the unfurling of the second lectin ring which enables tetramer formation. Furthermore, we suggest the relevance of this oligomerization pathway within a cellular setting, whereby cytosine-guanosine binding appeared to disrupt this cell-surface oligomer. Accordingly, we provide insight into the structure and oligomeric assembly of the DEC-205 receptor.
- Infection and Immunity Program, Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia; Australian Research Council Centre of Excellence for Advanced Molecular Imaging, Monash University, Clayton, Victoria, Australia. Electronic address: ben.gully@monash.edu.
Organizational Affiliation: