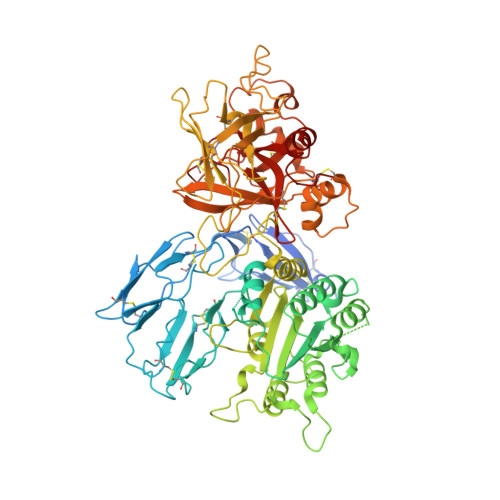
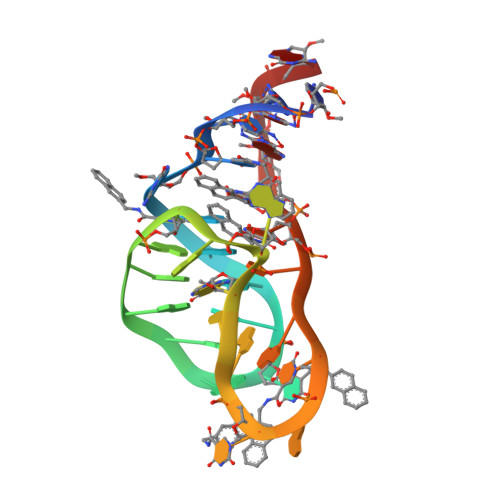
Inhibition of the Complement Alternative Pathway by Chemically Modified DNA Aptamers That Bind with Picomolar Affinity to Factor B.
Xu, X., Zhang, C., Denton, D.T., O'Connell, D., Drolet, D.W., Geisbrecht, B.V.(2021) J Immunol 206: 861-873
- PubMed: 33419768
- DOI: https://doi.org/10.4049/jimmunol.2001260
- Primary Citation of Related Structures:
7JTN, 7JTQ - PubMed Abstract:
The complement system is a conserved component of innate immunity that fulfills diverse roles in defense and homeostasis. Inappropriate activation of complement contributes to many inflammatory diseases, however, which has led to a renewed emphasis on development of therapeutic complement inhibitors. Activation of complement component C3 is required for amplification of complement and is achieved through two multisubunit proteases called C3 convertases. Of these, the alternative pathway (AP) C3 convertase is responsible for a majority of the C3 activation products in vivo, which renders it an attractive target for inhibitor discovery. In this study, we report the identification and characterization of two related slow off-rate modified DNA aptamers (SOMAmer) reagents that inhibit formation of the AP C3 convertase by binding to the proprotease, factor B (FB). These aptamers, known as SL1102 (31 bases) and SL1103 (29 bases), contain uniform substitutions of 5-( N -2-naphthylethylcarboxyamide)-2'-deoxyuridine for deoxythymidine. SL1102 and SL1103 bind FB with K d values of 49 and 88 pM, respectively, and inhibit activation of C3 and lysis of rabbit erythrocytes under AP-specific conditions. Cocrystal structures of SL1102 (3.4 Å) and SL1103 (3.1 Å) bound to human FB revealed that SL1102 and SL1103 recognize a site at the juncture of the CCP1, CCP3, and vWF domains of FB. Consistent with these structures and previously published information, these aptamers inhibited FB binding to C3b and blocked formation of the AP C3 convertase. Together, these results demonstrate potent AP inhibition by modified DNA aptamers and expand the pipeline of FB-binding molecules with favorable pharmacologic properties.
- Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, KS 66506.
Organizational Affiliation: