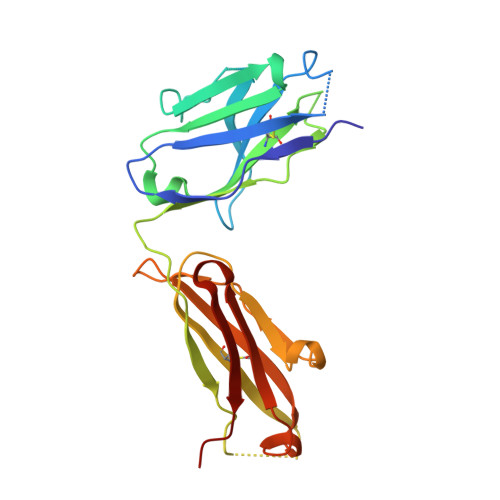
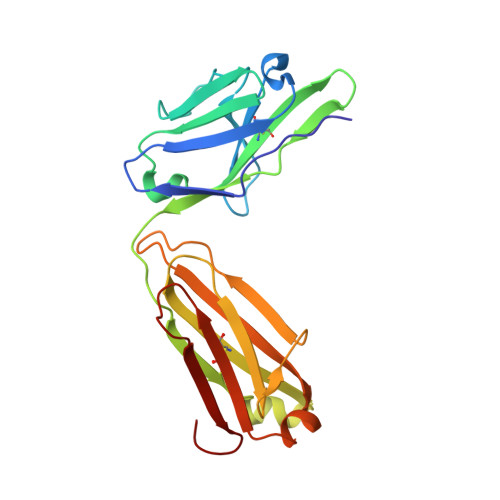
Potent SARS-CoV-2 binding and neutralization through maturation of iconic SARS-CoV-1 antibodies.
Rouet, R., Mazigi, O., Walker, G.J., Langley, D.B., Sobti, M., Schofield, P., Lenthall, H., Jackson, J., Ubiparipovic, S., Henry, J.Y., Abayasingam, A., Burnett, D., Kelleher, A., Brink, R., Bull, R.A., Turville, S., Stewart, A.G., Goodnow, C.C., Rawlinson, W.D., Christ, D.(null) MAbs 13: 1922134-1922134
- PubMed: 34024246
- DOI: https://doi.org/10.1080/19420862.2021.1922134
- Primary Citation of Related Structures:
7KZA, 7KZB, 7KZC - PubMed Abstract:
Antibodies against coronavirus spike protein potently protect against infection and disease, but whether such protection can be extended to variant coronaviruses is unclear. This is exemplified by a set of iconic and well-characterized monoclonal antibodies developed after the 2003 SARS outbreak, including mAbs m396, CR3022, CR3014 and 80R, which potently neutralize SARS-CoV-1, but not SARS-CoV-2. Here, we explore antibody engineering strategies to change and broaden their specificity, enabling nanomolar binding and potent neutralization of SARS-CoV-2. Intriguingly, while many of the matured clones maintained specificity of the parental antibody, new specificities were also observed, which was further confirmed by X-ray crystallography and cryo-electron microscopy, indicating that a limited set of VH antibody domains can give rise to variants targeting diverse epitopes, when paired with a diverse VL repertoire. Our findings open up over 15 years of antibody development efforts against SARS-CoV-1 to the SARS-CoV-2 field and outline general principles for the maturation of antibody specificity against emerging viruses.
- Immunology Department, Garvan Institute of Medical Research, Sydney, NSW, Australia.
Organizational Affiliation: