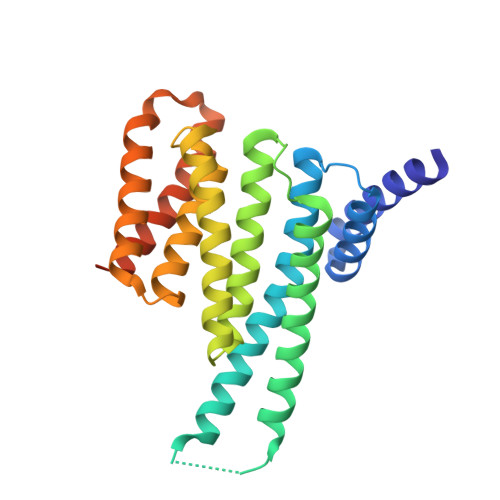
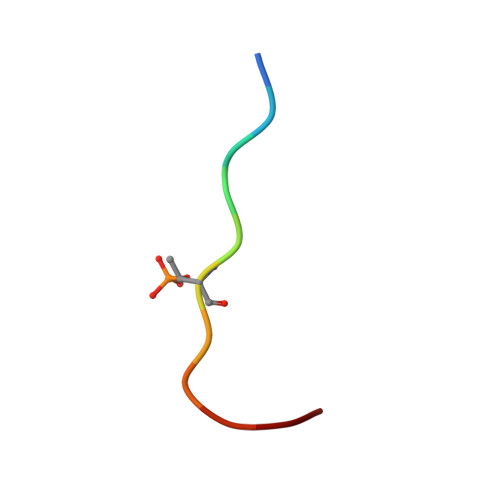
Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
Cossar, P.J., Wolter, M., van Dijck, L., Valenti, D., Levy, L.M., Ottmann, C., Brunsveld, L.(2021) J Am Chem Soc 143: 8454-8464
- PubMed: 34047554
- DOI: https://doi.org/10.1021/jacs.1c03035
- Primary Citation of Related Structures:
7AOG, 7AXN, 7AYF, 7AZ1, 7AZ2, 7BDP, 7BDT, 7BDY, 7BFW, 7BG3, 7BGQ, 7BGR, 7BGV, 7BGW, 7NIF, 7NIG, 7NIX, 7NJ6, 7NJ8, 7NJA, 7NQP, 7NRK, 7NRL, 7NSV - PubMed Abstract:
The stabilization of protein complexes has emerged as a promising modality, expanding the number of entry points for novel therapeutic intervention. Targeting proteins that mediate protein-protein interactions (PPIs), such as hub proteins, is equally challenging and rewarding as they offer an intervention platform for a variety of diseases, due to their large interactome. 14-3-3 hub proteins bind phosphorylated motifs of their interaction partners in a conserved binding channel. The 14-3-3 PPI interface is consequently only diversified by its different interaction partners. Therefore, it is essential to consider, additionally to the potency, also the selectivity of stabilizer molecules. Targeting a lysine residue at the interface of the composite 14-3-3 complex, which can be targeted explicitly via aldimine-forming fragments, we studied the de novo design of PPI stabilizers under consideration of potential selectivity. By applying cooperativity analysis of ternary complex formation, we developed a reversible covalent molecular glue for the 14-3-3/Pin1 interaction. This small fragment led to a more than 250-fold stabilization of the 14-3-3/Pin1 interaction by selective interfacing with a unique tryptophan in Pin1. This study illustrates how cooperative complex formation drives selective PPI stabilization. Further, it highlights how specific interactions within a hub proteins interactome can be stabilized over other interactions with a common binding motif.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands.
Organizational Affiliation: