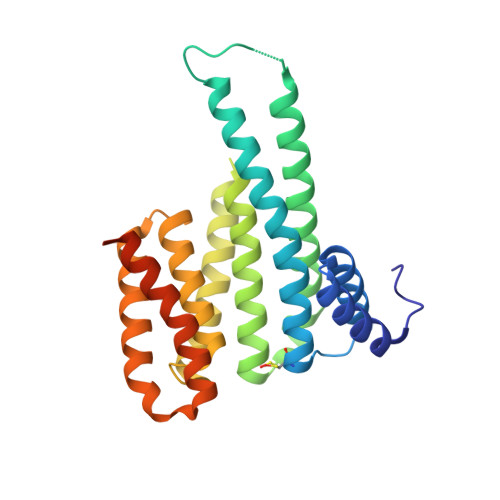
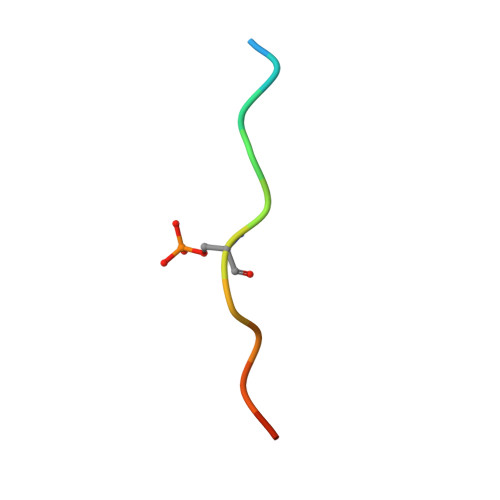
Fragment-based exploration of the 14-3-3/Amot-p130 interface.
Centorrino, F., Andlovic, B., Cossar, P., Brunsveld, L., Ottmann, C.(2022) Curr Res Struct Biol 4: 21-28
- PubMed: 35036934
- DOI: https://doi.org/10.1016/j.crstbi.2021.12.003
- Primary Citation of Related Structures:
7NMA, 7NMW, 7NMX, 7NN2, 7NND, 7NNE, 7NP2, 7NPB, 7NPG - PubMed Abstract:
The modulation of protein-protein interactions (PPIs) has developed into a well-established field of drug discovery. Despite the advances achieved in the field, many PPIs are still deemed as ' undruggable' targets and the design of PPIs stabilizers remains a significant challenge. The application of fragment-based methods for the identification of drug leads and to evaluate the ' tractability' of the desired protein target has seen a remarkable development in recent years. In this study, we explore the molecular characteristics of the 14-3-3/Amot-p130 PPI and the conceptual possibility of targeting this interface using X-ray crystallography fragment-based screening. We report the first structural elucidation of the 14-3-3 binding motif of Amot-p130 and the characterization of the binding mode and affinities involved. We made use of fragments to probe the ' ligandability' of the 14-3-3/Amot-p130 composite binding pocket. Here we disclose initial hits with promising stabilizing activity and an early-stage selectivity toward the Amot-p130 motifs over other representatives 14-3-3 partners. Our findings highlight the potential of using fragments to characterize and explore proteins' surfaces and might provide a starting point toward the development of small molecules capable of acting as molecular glues .
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, P. O. Box 513, 5600 MB, Eindhoven, the Netherlands.
Organizational Affiliation: