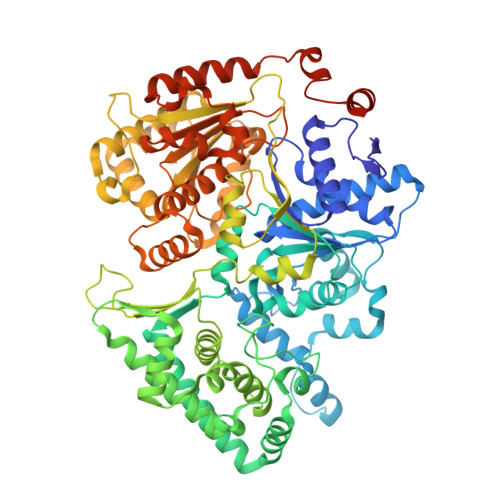
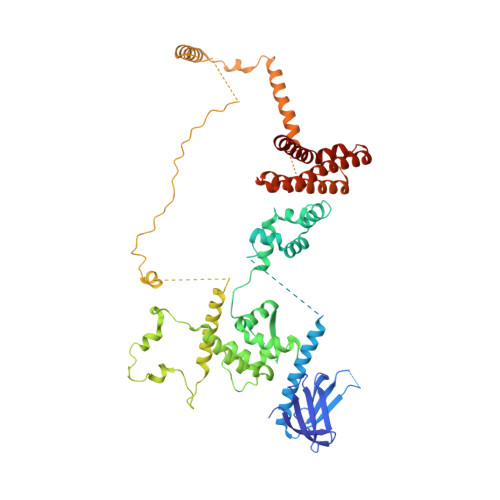
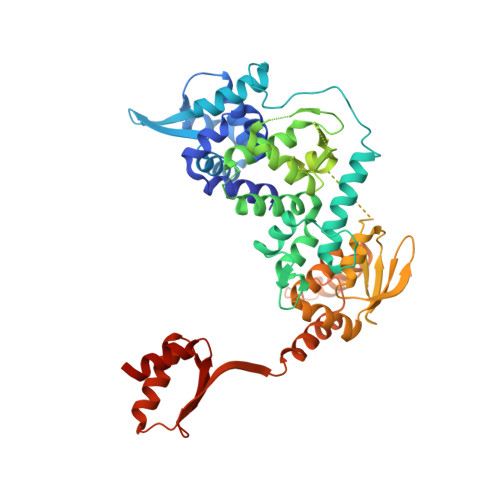
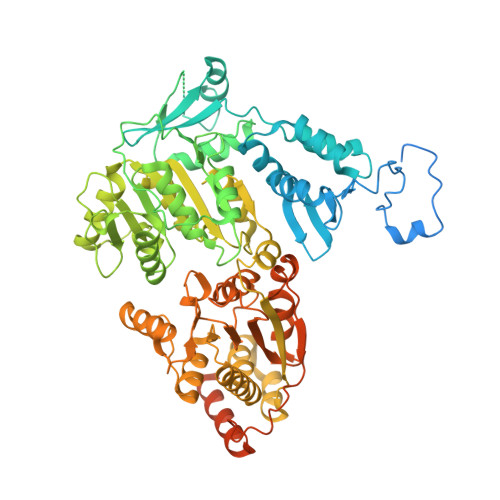
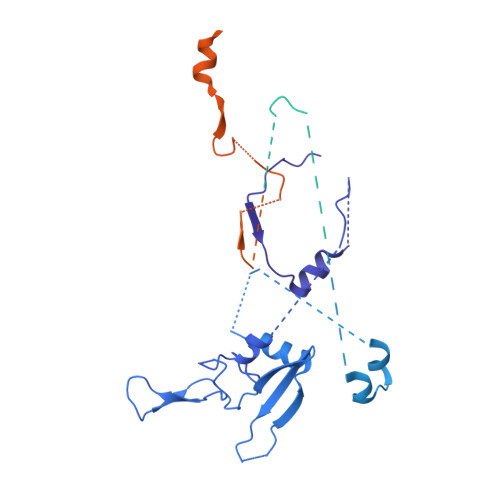
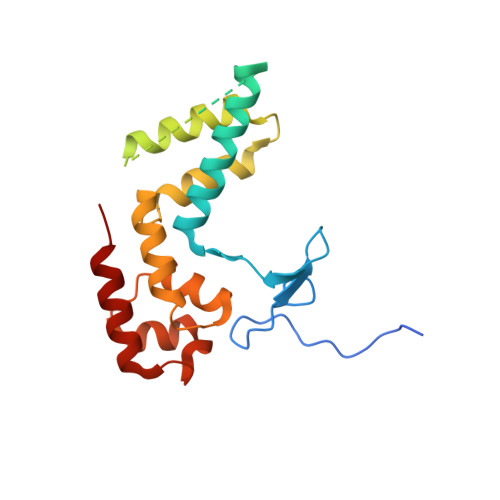
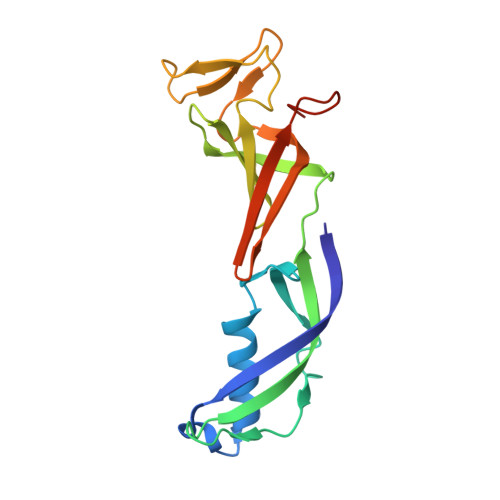
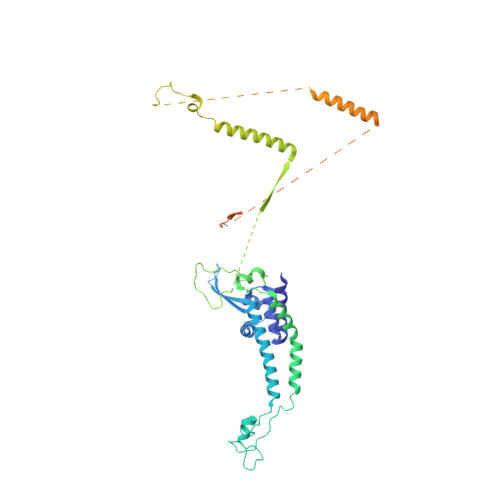
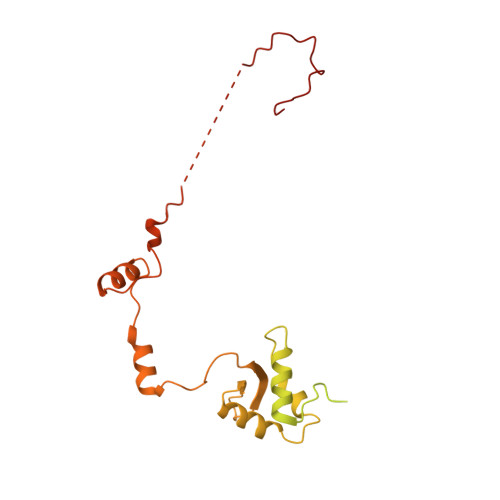
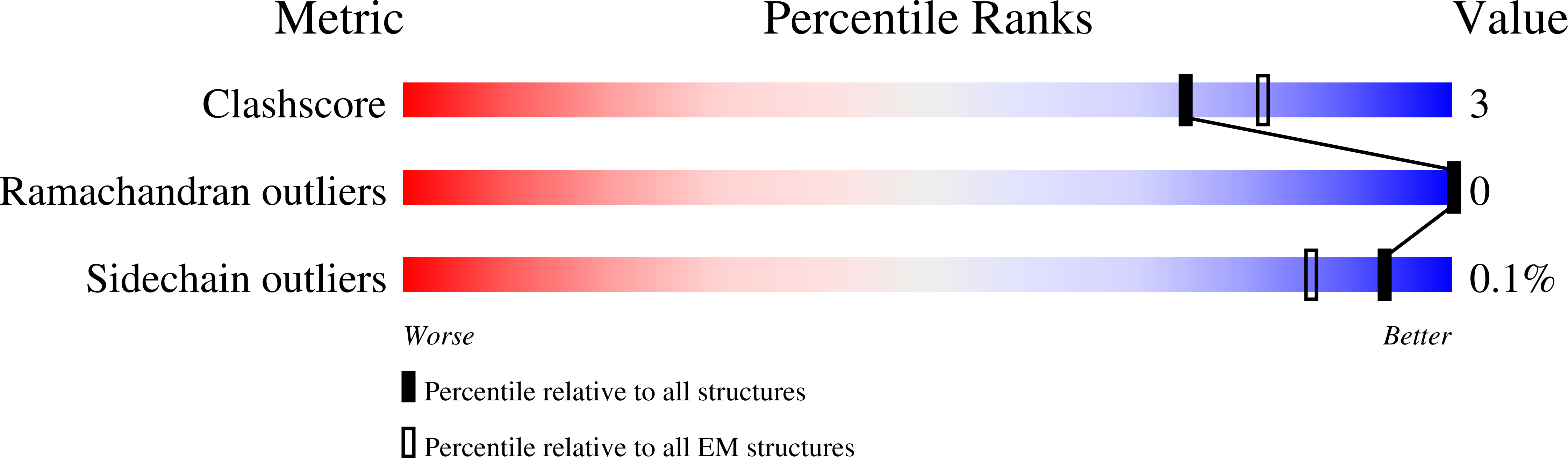Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Schilbach, S., Aibara, S., Dienemann, C., Grabbe, F., Cramer, P.(2021) Cell 184: 4064-4072.e28
- PubMed: 34133942
- DOI: https://doi.org/10.1016/j.cell.2021.05.012
- Primary Citation of Related Structures:
7O4I, 7O4J, 7O4K, 7O4L, 7O72, 7O73, 7O75 - PubMed Abstract:
Transcription initiation requires assembly of the RNA polymerase II (Pol II) pre-initiation complex (PIC) and opening of promoter DNA. Here, we present the long-sought high-resolution structure of the yeast PIC and define the mechanism of initial DNA opening. We trap the PIC in an intermediate state that contains half a turn of open DNA located 30-35 base pairs downstream of the TATA box. The initially opened DNA region is flanked and stabilized by the polymerase "clamp head loop" and the TFIIF "charged region" that both contribute to promoter-initiated transcription. TFIIE facilitates initiation by buttressing the clamp head loop and by regulating the TFIIH translocase. The initial DNA bubble is then extended in the upstream direction, leading to the open promoter complex and enabling start-site scanning and RNA synthesis. This unique mechanism of DNA opening may permit more intricate regulation than in the Pol I and Pol III systems.
Organizational Affiliation:
Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.