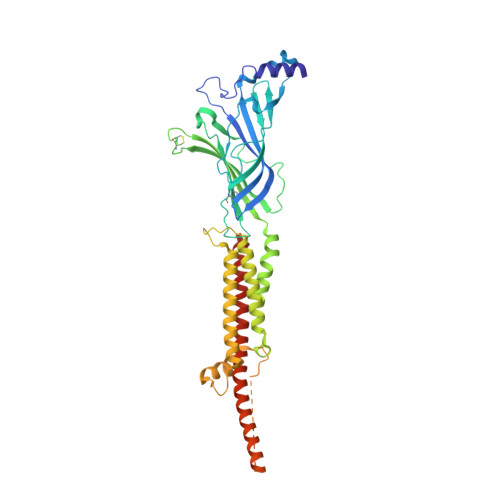
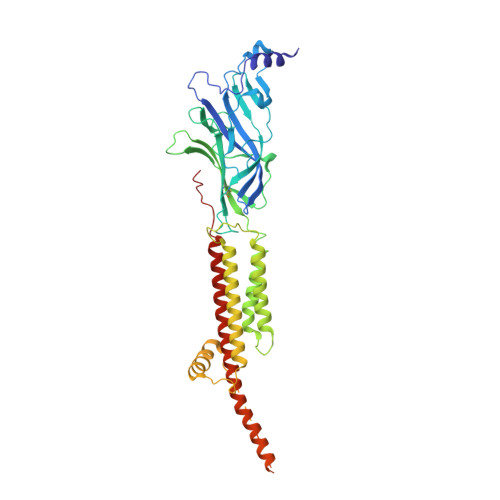
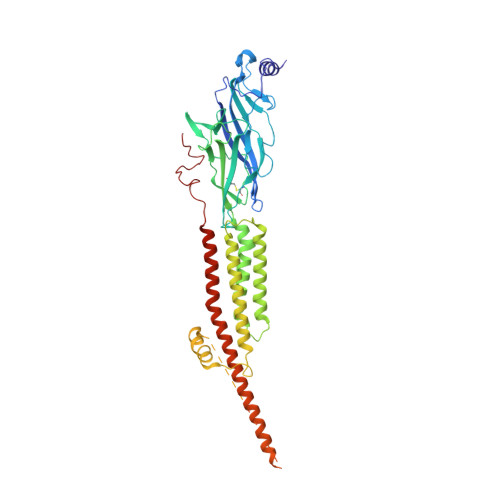
Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Zarkadas, E., Pebay-Peyroula, E., Thompson, M.J., Schoehn, G., Uchanski, T., Steyaert, J., Chipot, C., Dehez, F., Baenziger, J.E., Nury, H.(2022) Neuron 110: 1358
- PubMed: 35139364
- DOI: https://doi.org/10.1016/j.neuron.2022.01.013
- Primary Citation of Related Structures:
7QKO, 7QL5, 7QL6 - PubMed Abstract:
Fast synaptic communication requires receptors that respond to the presence of neurotransmitter by opening an ion channel across the post-synaptic membrane. The muscle-type nicotinic acetylcholine receptor from the electric fish, Torpedo, is the prototypic ligand-gated ion channel, yet the structural changes underlying channel activation remain undefined. Here we use cryo-EM to solve apo and agonist-bound structures of the Torpedo nicotinic receptor embedded in a lipid nanodisc. Using both a direct biochemical assay to define the conformational landscape and molecular dynamics simulations to assay flux through the pore, we correlate structures with functional states and elucidate the motions that lead to pore activation of a heteromeric nicotinic receptor. We highlight an underappreciated role for the complementary subunit in channel gating, establish the structural basis for the differential agonist affinities of α/δ versus α /γ sites, and explain why nicotine is less potent at muscle nicotinic receptors compared to neuronal ones.
Organizational Affiliation:
Université Grenoble Alpes, CNRS, CEA, IBS, F-38000 Grenoble, France.