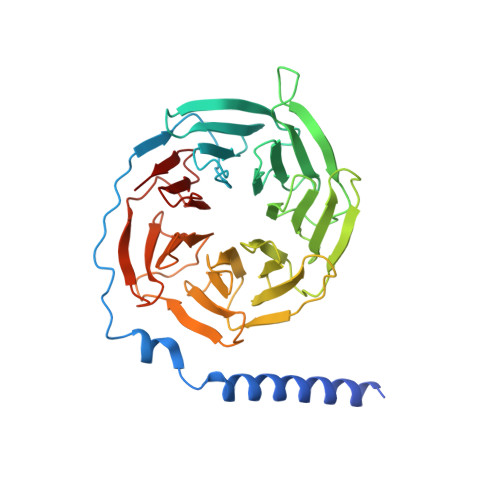
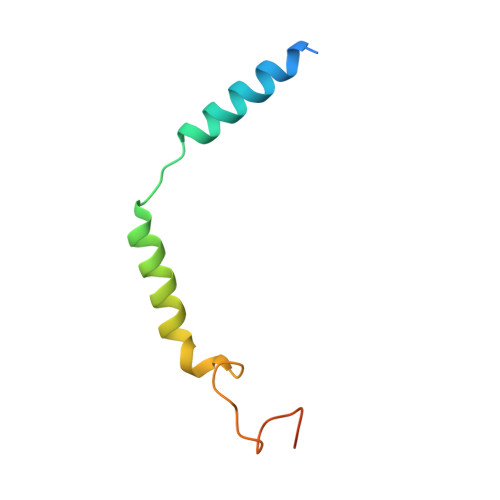
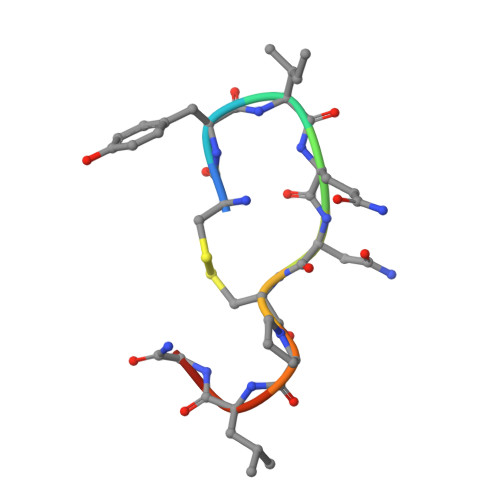
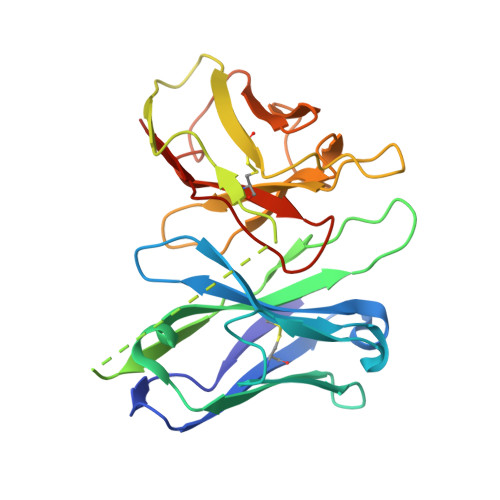
Structural basis for the activation and ligand recognition of the human oxytocin receptor.
Waltenspuhl, Y., Ehrenmann, J., Vacca, S., Thom, C., Medalia, O., Pluckthun, A.(2022) Nat Commun 13: 4153-4153
- PubMed: 35851571
- DOI: https://doi.org/10.1038/s41467-022-31325-0
- Primary Citation of Related Structures:
7QVM - PubMed Abstract:
The small cyclic neuropeptide hormone oxytocin (OT) and its cognate receptor play a central role in the regulation of social behaviour and sexual reproduction. Here we report the single-particle cryo-electron microscopy structure of the active oxytocin receptor (OTR) in complex with its cognate ligand oxytocin. Our structure provides high-resolution insights into the OT binding mode, the OTR activation mechanism as well as the subtype specificity within the oxytocin/vasopressin receptor family.
Organizational Affiliation:
Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057, Zürich, Switzerland.