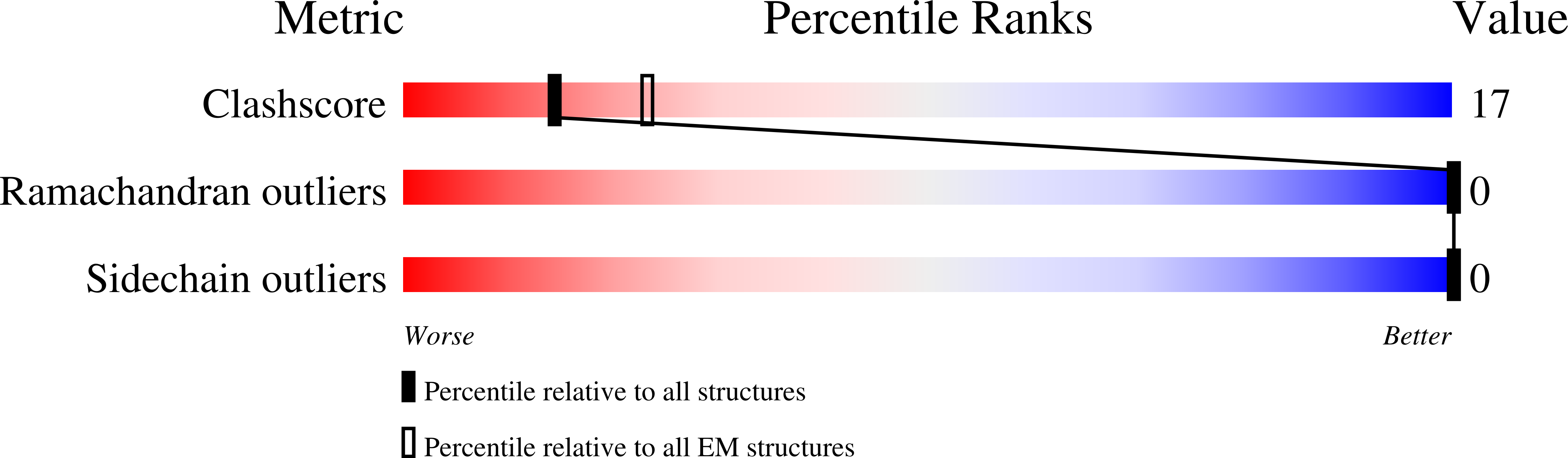Structure of a nucleosome-bound MuvB transcription factor complex reveals DNA remodelling.
Koliopoulos, M.G., Muhammad, R., Roumeliotis, T.I., Beuron, F., Choudhary, J.S., Alfieri, C.(2022) Nat Commun 13: 5075-5075
- PubMed: 36038598
- DOI: https://doi.org/10.1038/s41467-022-32798-9
- Primary Citation of Related Structures:
7R1D - PubMed Abstract:
Genes encoding the core cell cycle machinery are transcriptionally regulated by the MuvB family of protein complexes in a cell cycle-specific manner. Complexes of MuvB with the transcription factors B-MYB and FOXM1 activate mitotic genes during cell proliferation. The mechanisms of transcriptional regulation by these complexes are still poorly characterised. Here, we combine biochemical analysis and in vitro reconstitution, with structural analysis by cryo-electron microscopy and cross-linking mass spectrometry, to functionally examine these complexes. We find that the MuvB:B-MYB complex binds and remodels nucleosomes, thereby exposing nucleosomal DNA. This remodelling activity is supported by B-MYB which directly binds the remodelled DNA. Given the remodelling activity on the nucleosome, we propose that the MuvB:B-MYB complex functions as a pioneer transcription factor complex. In this work, we rationalise prior biochemical and cellular studies and provide a molecular framework of interactions on a protein complex that is key for cell cycle regulation.
Organizational Affiliation:
Division of Structural Biology, Chester Beatty Laboratories, The Institute of Cancer Research, London, UK.