AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Mosalaganti, S., Obarska-Kosinska, A., Siggel, M., Taniguchi, R., Turonova, B., Zimmerli, C.E., Buczak, K., Schmidt, F.H., Margiotta, E., Mackmull, M.T., Hagen, W.J.H., Hummer, G., Kosinski, J., Beck, M.(2022) Science 376: eabm9506-eabm9506
- PubMed: 35679397
- DOI: https://doi.org/10.1126/science.abm9506
- Primary Citation of Related Structures:
7R1Y, 7R5J, 7R5K - PubMed Abstract:
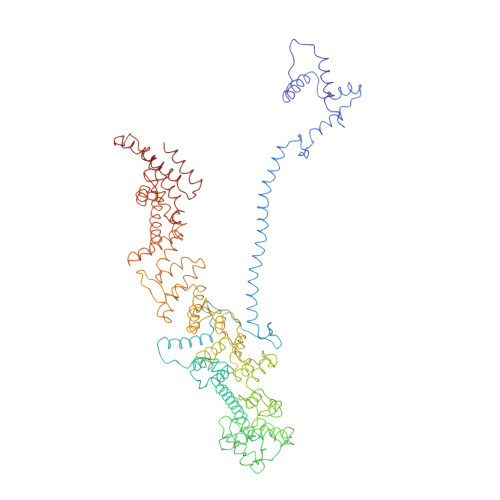
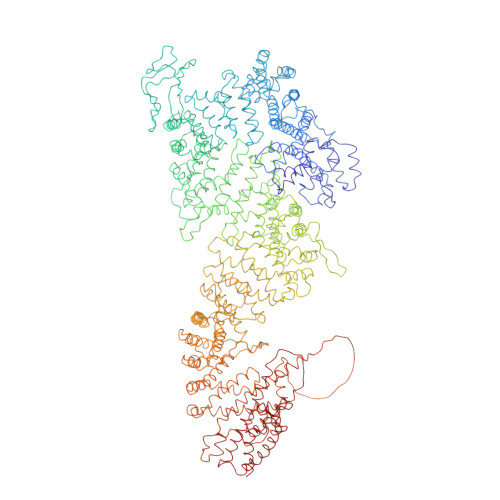
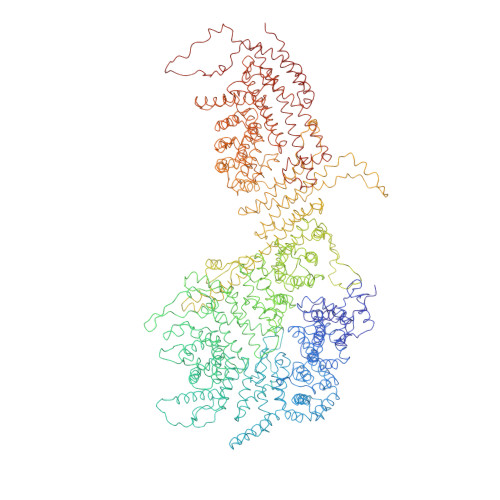
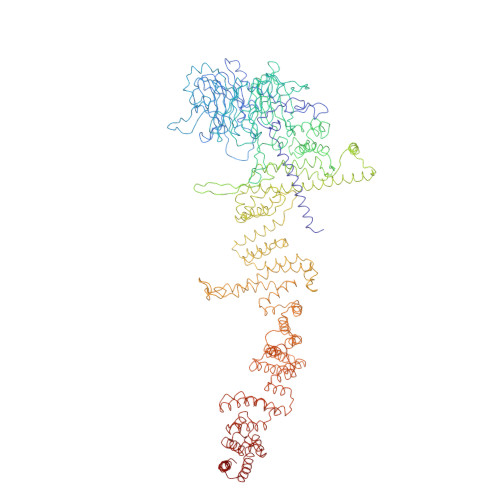
INTRODUCTION The eukaryotic nucleus pro-tects the genome and is enclosed by the two membranes of the nuclear envelope. Nuclear pore complexes (NPCs) perforate the nuclear envelope to facilitate nucleocytoplasmic transport. With a molecular weight of ∼120 MDa, the human NPC is one of the larg-est protein complexes. Its ~1000 proteins are taken in multiple copies from a set of about 30 distinct nucleoporins (NUPs). They can be roughly categorized into two classes. Scaf-fold NUPs contain folded domains and form a cylindrical scaffold architecture around a central channel. Intrinsically disordered NUPs line the scaffold and extend into the central channel, where they interact with cargo complexes. The NPC architecture is highly dynamic. It responds to changes in nuclear envelope tension with conforma-tional breathing that manifests in dilation and constriction movements. Elucidating the scaffold architecture, ultimately at atomic resolution, will be important for gaining a more precise understanding of NPC function and dynamics but imposes a substantial chal-lenge for structural biologists. RATIONALE Considerable progress has been made toward this goal by a joint effort in the field. A synergistic combination of complementary approaches has turned out to be critical. In situ structural biology techniques were used to reveal the overall layout of the NPC scaffold that defines the spatial reference for molecular modeling. High-resolution structures of many NUPs were determined in vitro. Proteomic analysis and extensive biochemical work unraveled the interaction network of NUPs. Integra-tive modeling has been used to combine the different types of data, resulting in a rough outline of the NPC scaffold. Previous struc-tural models of the human NPC, however, were patchy and limited in accuracy owing to several challenges: (i) Many of the high-resolution structures of individual NUPs have been solved from distantly related species and, consequently, do not comprehensively cover their human counterparts. (ii) The scaf-fold is interconnected by a set of intrinsically disordered linker NUPs that are not straight-forwardly accessible to common structural biology techniques. (iii) The NPC scaffold intimately embraces the fused inner and outer nuclear membranes in a distinctive topol-ogy and cannot be studied in isolation. (iv) The conformational dynamics of scaffold NUPs limits the resolution achievable in structure determination. RESULTS In this study, we used artificial intelligence (AI)-based prediction to generate an exten-sive repertoire of structural models of human NUPs and their subcomplexes. The resulting models cover various domains and interfaces that so far remained structurally uncharac-terized. Benchmarking against previous and unpublished x-ray and cryo-electron micros-copy structures revealed unprecedented accu-racy. We obtained well-resolved cryo-electron tomographic maps of both the constricted and dilated conformational states of the hu-man NPC. Using integrative modeling, we fit-ted the structural models of individual NUPs into the cryo-electron microscopy maps. We explicitly included several linker NUPs and traced their trajectory through the NPC scaf-fold. We elucidated in great detail how mem-brane-associated and transmembrane NUPs are distributed across the fusion topology of both nuclear membranes. The resulting architectural model increases the structural coverage of the human NPC scaffold by about twofold. We extensively validated our model against both earlier and new experimental data. The completeness of our model has enabled microsecond-long coarse-grained molecular dynamics simulations of the NPC scaffold within an explicit membrane en-vironment and solvent. These simulations reveal that the NPC scaffold prevents the constriction of the otherwise stable double-membrane fusion pore to small diameters in the absence of membrane tension. CONCLUSION Our 70-MDa atomically re-solved model covers >90% of the human NPC scaffold. It captures conforma-tional changes that occur during dilation and constriction. It also reveals the precise anchoring sites for intrinsically disordered NUPs, the identification of which is a prerequisite for a complete and dy-namic model of the NPC. Our study exempli-fies how AI-based structure prediction may accelerate the elucidation of subcellular ar-chitecture at atomic resolution. [Figure: see text].
Organizational Affiliation:
Department of Molecular Sociology, Max Planck Institute of Biophysics, 60438 Frankfurt am Main, Germany.