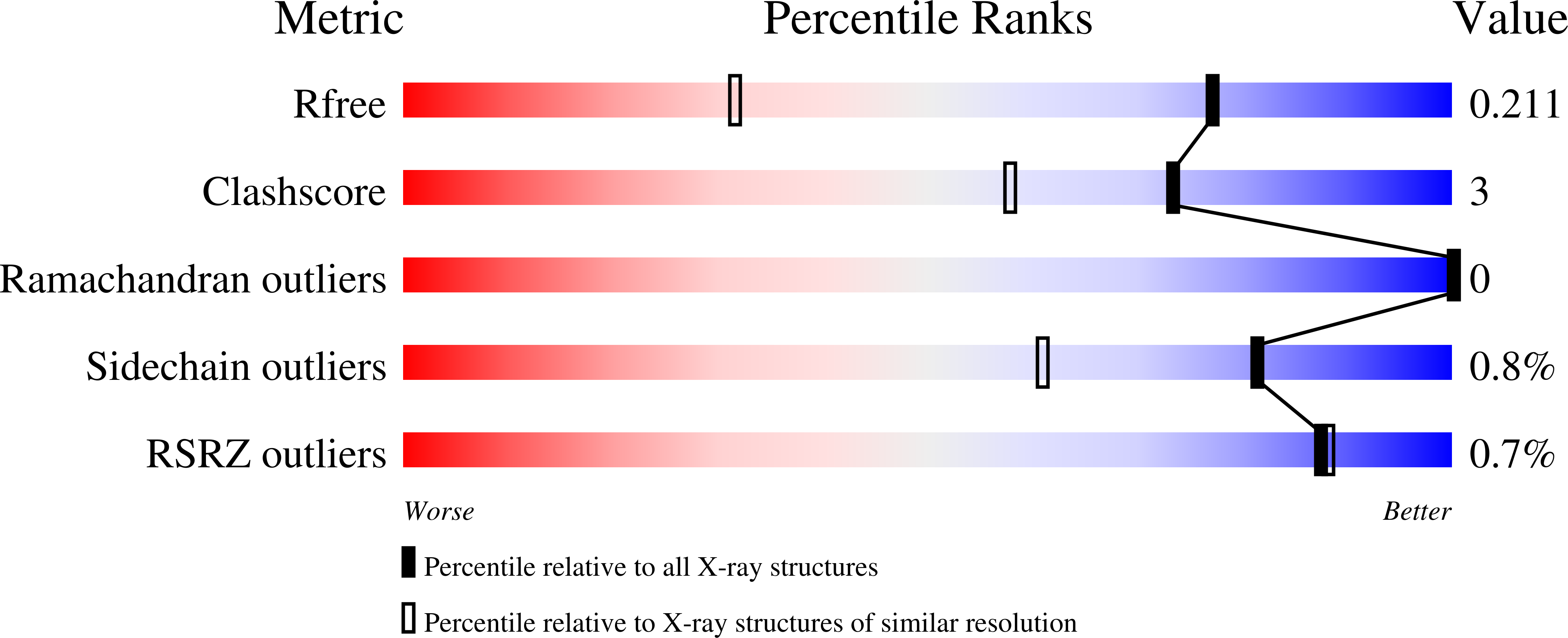
A two-directional vibrational probe reveals different electric field orientations in solution and an enzyme active site.
Zheng, C., Mao, Y., Kozuch, J., Atsango, A.O., Ji, Z., Markland, T.E., Boxer, S.G.(2022) Nat Chem 14: 891-897
- PubMed: 35513508
- DOI: https://doi.org/10.1038/s41557-022-00937-w
- Primary Citation of Related Structures:
7RM6 - PubMed Abstract:
The catalytic power of an electric field depends on its magnitude and orientation with respect to the reactive chemical species. Understanding and designing new catalysts for electrostatic catalysis thus requires methods to measure the electric field orientation and magnitude at the molecular scale. We demonstrate that electric field orientations can be extracted using a two-directional vibrational probe by exploiting the vibrational Stark effect of both the C=O and C-D stretches of a deuterated aldehyde. Combining spectroscopy with molecular dynamics and electronic structure partitioning methods, we demonstrate that, despite distinct polarities, solvents act similarly in their preference for electrostatically stabilizing large bond dipoles at the expense of destabilizing small ones. In contrast, we find that for an active-site aldehyde inhibitor of liver alcohol dehydrogenase, the electric field orientation deviates markedly from that found in solvents, which provides direct evidence for the fundamental difference between the electrostatic environment of solvents and that of a preorganized enzyme active site.
Organizational Affiliation:
Department of Chemistry, Stanford University, Stanford, CA, USA.