Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation.
Ma, M.T., Jennings, M.R., Blazeck, J., Lieberman, R.L.(2022) Acta Crystallogr D Struct Biol 78: 91-103
- PubMed: 34981765
- DOI: https://doi.org/10.1107/S2059798321011785
- Primary Citation of Related Structures:
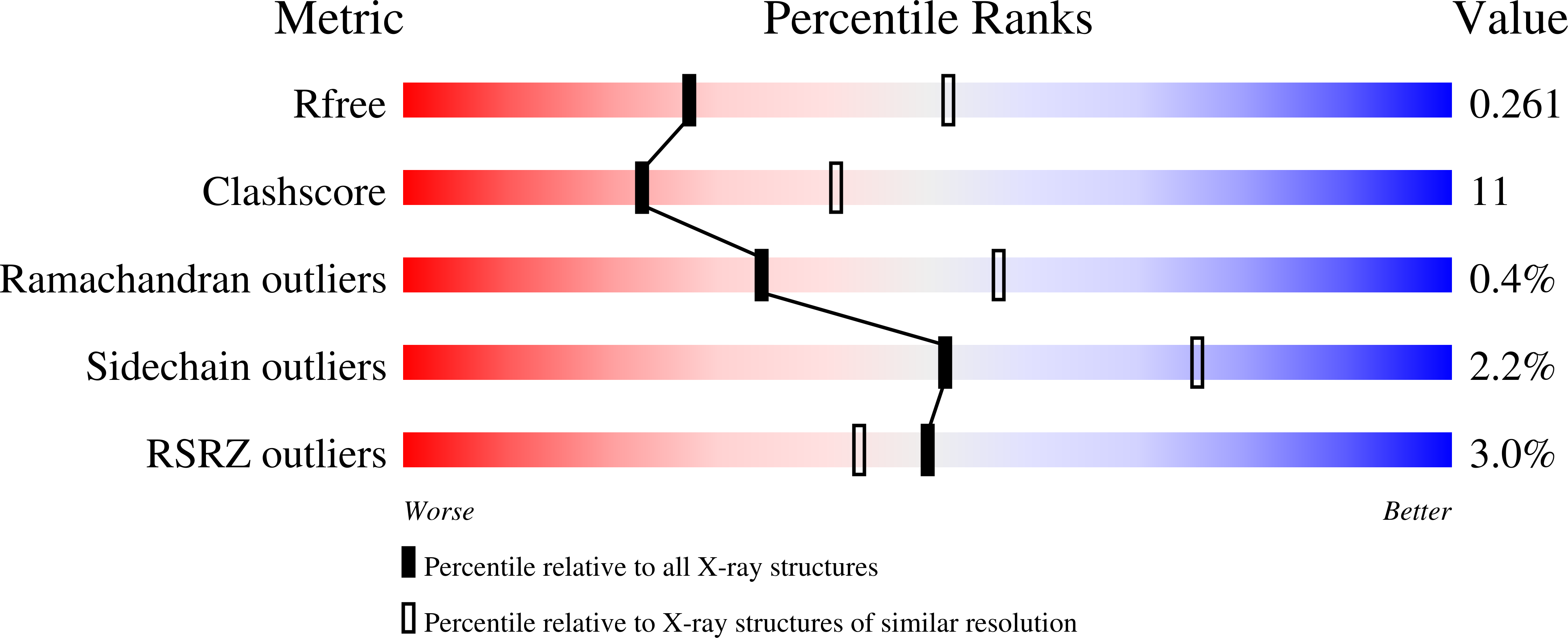
7RTG - PubMed Abstract:
Homo sapiens adenosine deaminase 1 (HsADA1; UniProt P00813) is an immunologically relevant enzyme with roles in T-cell activation and modulation of adenosine metabolism and signaling. Patients with genetic deficiency in HsADA1 suffer from severe combined immunodeficiency, and HsADA1 is a therapeutic target in hairy cell leukemias. Historically, insights into the catalytic mechanism and the structural attributes of HsADA1 have been derived from studies of its homologs from Bos taurus (BtADA) and Mus musculus (MmADA). Here, the structure of holo HsADA1 is presented, as well as biochemical characterization that confirms its high activity and shows that it is active across a broad pH range. Structurally, holo HsADA1 adopts a closed conformation distinct from the open conformation of holo BtADA. Comparison of holo HsADA1 and MmADA reveals that MmADA also adopts a closed conformation. These findings challenge previous assumptions gleaned from BtADA regarding the conformation of HsADA1 that may be relevant to its immunological interactions, particularly its ability to bind adenosine receptors. From a broader perspective, the structural analysis of HsADA1 presents a cautionary tale for reliance on homologs to make structural inferences relevant to applications such as protein engineering or drug development.
Organizational Affiliation:
School of Chemistry and Biochemistry, Georgia Institute of Technology, 901 Atlantic Drive NW, Atlanta, GA 30332, USA.