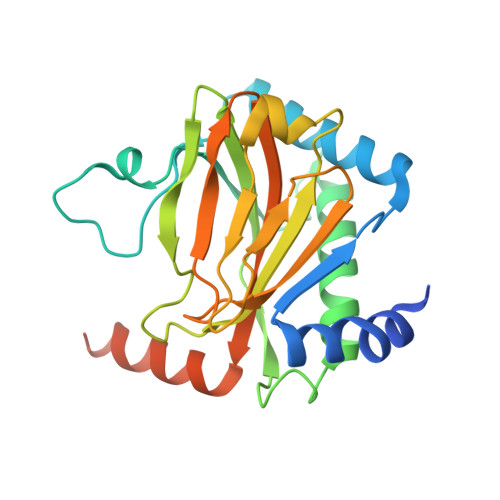
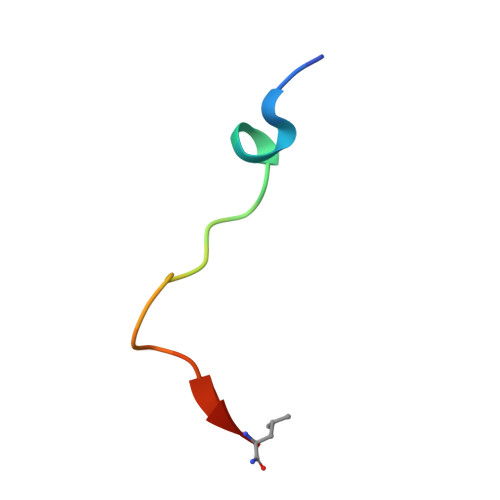
Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Ferens, F.G., Taber, C.C., Stuart, S., Hubert, M., Tarade, D., Lee, J.E., Ohh, M.(2024) Commun Biol 7: 240-240
- PubMed: 38418569
- DOI: https://doi.org/10.1038/s42003-024-05904-4
- Primary Citation of Related Structures:
7UJV - PubMed Abstract:
Pacak-Zhuang syndrome is caused by mutations in the EPAS1 gene, which encodes for one of the three hypoxia-inducible factor alpha (HIFα) paralogs HIF2α and is associated with defined but varied phenotypic presentations including neuroendocrine tumors and polycythemia. However, the mechanisms underlying the complex genotype-phenotype correlations remain incompletely understood. Here, we devised a quantitative method for determining the dissociation constant (K d ) of the HIF2α peptides containing disease-associated mutations and the catalytic domain of prolyl-hydroxylase (PHD2) using microscale thermophoresis (MST) and showed that neuroendocrine-associated Class 1 HIF2α mutants have distinctly higher K d than the exclusively polycythemia-associated Class 2 HIF2α mutants. Based on the co-crystal structure of PHD2/HIF2α peptide complex at 1.8 Å resolution, we showed that the Class 1 mutated residues are localized to the critical interface between HIF2α and PHD2, adjacent to the PHD2 active catalytic site, while Class 2 mutated residues are localized to the more flexible region of HIF2α that makes less contact with PHD2. Concordantly, Class 1 mutations were found to significantly increase HIF2α-mediated transcriptional activation in cellulo compared to Class 2 counterparts. These results reveal a structural mechanism in which the strength of the interaction between HIF2α and PHD2 is at the root of the general genotype-phenotype correlations observed in Pacak-Zhuang syndrome.
Organizational Affiliation:
Department of Laboratory Medicine & Pathobiology, Faculty of Medicine, University of Toronto, 1 King's College Circle, Toronto, ON, M5S 1A8, Canada.