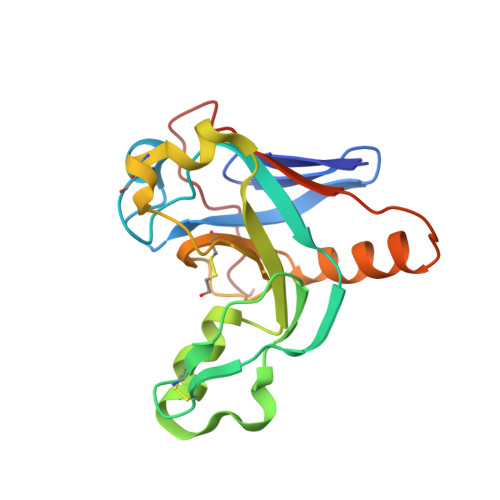
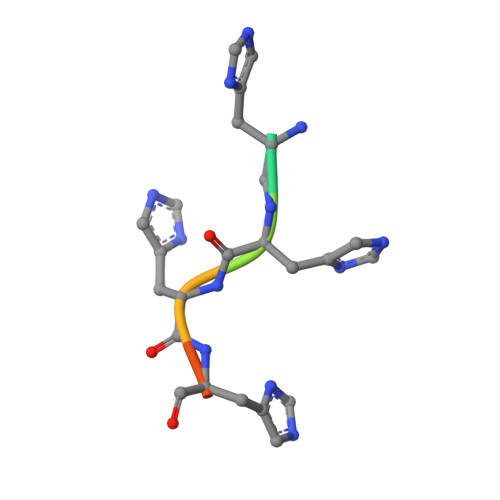
Molecular characterization of the type VI secretion system effector Tlde1a reveals a structurally altered LD-transpeptidase fold.
Lorente Cobo, N., Sibinelli-Sousa, S., Biboy, J., Vollmer, W., Bayer-Santos, E., Prehna, G.(2022) J Biological Chem 298: 102556-102556
- PubMed: 36183829
- DOI: https://doi.org/10.1016/j.jbc.2022.102556
- Primary Citation of Related Structures:
7UMA, 7UO3, 7UO8 - PubMed Abstract:
The type VI secretion system (T6SS) is a molecular machine that Gram-negative bacteria have adapted for multiple functions, including interbacterial competition. Bacteria use the T6SS to deliver protein effectors into adjacent cells to kill rivals and establish niche dominance. Central to T6SS-mediated bacterial competition is an arms race to acquire diverse effectors to attack and neutralize target cells. The peptidoglycan has a central role in bacterial cell physiology, and effectors that biochemically modify peptidoglycan structure effectively induce cell death. One such T6SS effector is Tlde1a from Salmonella Typhimurium. Tlde1a functions as an LD-carboxypeptidase to cleave tetrapeptide stems and as an LD-transpeptidase to exchange the terminal D-alanine of a tetrapeptide stem with a noncanonical D-amino acid. To understand how Tlde1a exhibits toxicity at the molecular level, we determined the X-ray crystal structure of Tlde1a alone and in complex with D-amino acids. Our structural data revealed that Tlde1a possesses a unique LD-transpeptidase fold consisting of a dual pocket active site with a capping subdomain. This includes an exchange pocket to bind a D-amino acid for exchange and a catalytic pocket to position the D-alanine of a tetrapeptide stem for cleavage. Our toxicity assays in Escherichia coli and in vitro peptidoglycan biochemical assays with Tlde1a variants correlate Tlde1a molecular features directly to its biochemical functions. We observe that the LD-carboxypeptidase and LD-transpeptidase activities of Tlde1a are both structurally and functionally linked. Overall, our data highlight how an LD-transpeptidase fold has been structurally altered to create a toxic effector in the T6SS arms race.
- Department of Microbiology, University of Manitoba, Winnipeg, Canada.
Organizational Affiliation: