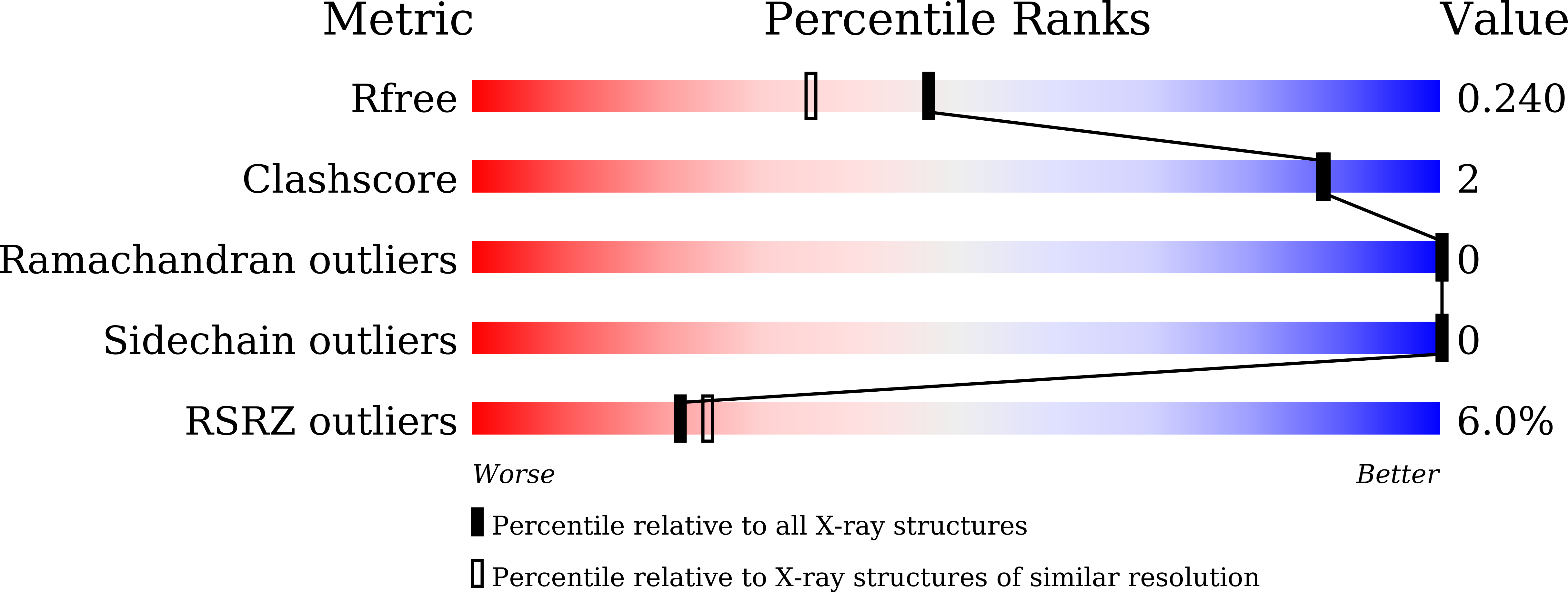
Structure-based design of a novel inhibitor of the ZIKA virus NS2B/NS3 protease.
Xiong, Y., Cheng, F., Zhang, J., Su, H., Hu, H., Zou, Y., Li, M., Xu, Y.(2022) Bioorg Chem 128: 106109-106109
- PubMed: 36049322
- DOI: https://doi.org/10.1016/j.bioorg.2022.106109
- Primary Citation of Related Structures:
7VXX, 7VXY - PubMed Abstract:
Zika virus (ZIKV) has been a serious public health problem, and there is no vaccine or drug approved for the prevention or treatment of ZIKV yet. The ZIKV NS2B/NS3 protease plays an important role in processing the virus precursor polyprotein and is thus a promising target for antiviral drugs development. In order to discover novel inhibitors of this protease, we carried out a fragment-based hit screening and characterized protein-inhibitor interactions using the X-ray crystallography together with isothermal titration calorimetry. We reported two high-resolution crystal structures of the protease (bZiPro C143S ) in complex with an active fragment as well as a tetrapeptide, revealing that there is domain swapping in the protein structures and two ligands only occupy the substrate-binding pocket of one copy in a symmetric unit. Based on the detailed binding modes of two ligands revealed by crystal structures, we designed a novel inhibitor which inhibits the NS2B/NS3 protease with a higher potency than the fragment and possesses a higher ligand-binding efficiency and a comparable IC 50 compared to the tetrapeptide. These results thus provide a structural basis and valuable hint for development of more potent inhibitors of the ZIKV NS2B/NS3 protease.
Organizational Affiliation:
CAS Key Laboratory of Receptor Research and State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; University of Chinese Academy of Sciences, Beijing 100049, China.