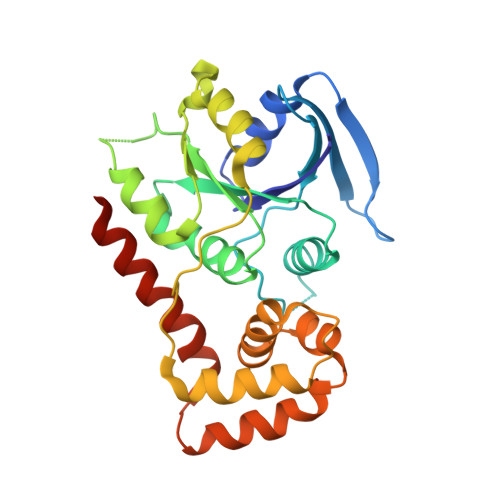
FeoC from Klebsiella pneumoniae uses its iron sulfur cluster to regulate the GTPase activity of the ferrous iron channel.
Hsueh, K.L., Yu, L.K., Hsieh, Y.C., Hsiao, Y.Y., Chen, C.J.(2023) Biochim Biophys Acta Proteins Proteom 1871: 140855-140855
- PubMed: 36182071
- DOI: https://doi.org/10.1016/j.bbapap.2022.140855
- Primary Citation of Related Structures:
7WQU - PubMed Abstract:
Bacteria depend on the ferrous iron transport (Feo) system for the uptake of ferrous iron (Fe 2+ ). The Feo system is crucial for colonization and virulence of pathogens. In γ-proteobacteria, the system consists of FeoA, FeoB, and FeoC. The function of FeoA remains unknown. FeoB likely forms the channel, whose regulation has been suggested to involve its GTPase domain (part of its NFeoB domain). FeoC from Klebsiella pneumonia was found to contain a [4Fe4S] cofactor, whose presence was speculated to enhance the GTPase activity of FeoB (Hsueh, K.-L., et al., J. Bacteriol. 2013 195(20): 4726-34). We present results here that support and extend that hypothesis. We monitored the GTPase activity of FeoB by NMR spectroscopy and found that the presence of 7% FeoC-[4Fe-4S] 3+ (the highest level of cofactor achieved in vitro) increased the GTPase rate of NFeoB by 3.6-fold over NFeoB. The effect depends on the oxidation state of the cluster; with reduction of the cluster to [4Fe-4S] 2+ the GTPase greatly decreased the GTPase rate. From the effects of point mutations in FeoC on GTPase rates, we conclude that Lys62 and Lys68 on FeoC each contribute to increased GTPase activity on NFeoB. Mutation of Thr37 of NFeoB to Ser nearly abolished the GTPase activity. The GTPase activity of the isolated K. pneumoniae NFeoB-FeoC complex (NFeoBC) was found to be higher in KCl than in NaCl solution. We solved the X-ray structure of the NFeoBC crystallized from KCl and compared it with a prior X-ray structure crystalized from NaCl. We propose a hypothesis, consistent with these results, to explain the factors that influence the GTPase activity. Bacteria may use the oxygen-sensitive cluster as a sensor to up-regulate the gate closing speed.
Organizational Affiliation:
Institute of Biomedical Sciences, Academia Sinica, Taipei 11574, Taiwan; Department of Animal Science and Biotechnology, Tunghai University, Taichung 407224, Taiwan. Electronic address: ren.shiue@gmail.com.